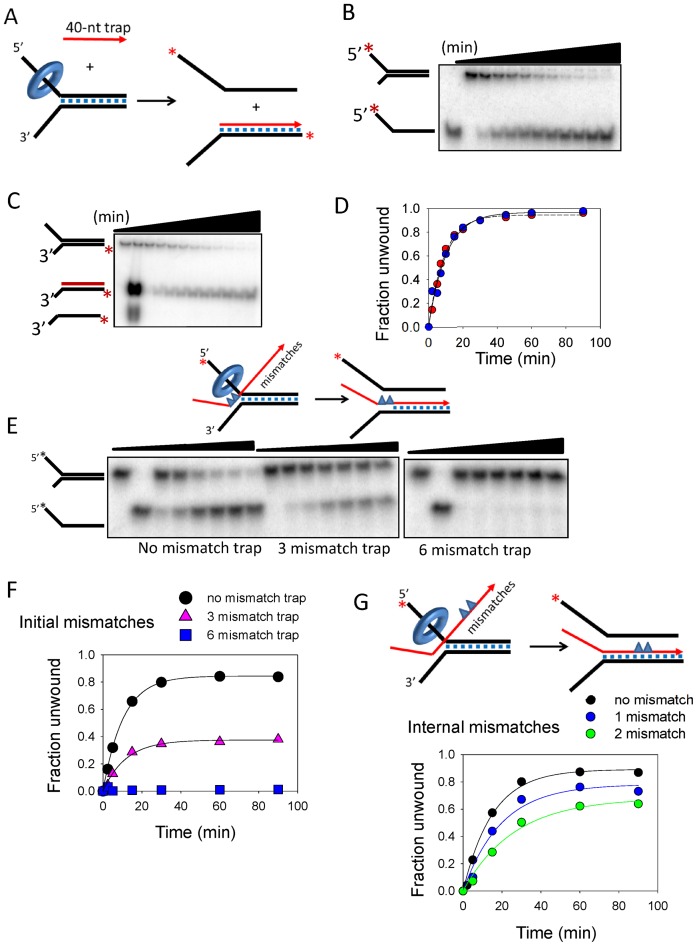
Figure 2.
TWINKLE catalyzed strand-exchange reaction is affected by mismatches in the recombining strands. (A) Experimental design to observe the strand-exchanged product using a displaced strand trap (40-nt trap) that is complementary only to the duplex region of the 3′-tail strand. (B) Gel image shows the time course of 5′-tail strand release from the 50% GC fork by TWINKLE in the presence of 200 nM 40-nt trap. (C) Gel image shows the time-dependent generation of the radiolabeled 3′-tail strand annealed to the 40-nt trap. (D) Quantitation of the gel images shows close match between the rates (0.09 ± 0.02 strand/min) of 5′-tail strand release in (B), and accumulation of the strand-exchanged product in (C). (E) Gel images show the time-dependent 5′-tail strand release by TWINKLE in the presence of ‘0 mismatch Trap’ (left image), ‘3-Ini mismatch Trap’ with three consecutive mismatches (middle) or ‘6-Ini mismatch Trap’ with six consecutive mismatches (right) in the initially unwound region at the fork junction, as shown in the cartoon. (F) Quantitation of the gel images in (E) shows the effect of mismatched traps on the kinetics of DNA unwinding. The initial rate of unwinding with no mismatched trap is 0.08 ± 0.01 strand/min and with three mismatched trap is 0.04 ± 0.01 strand/min. (G) Kinetics of 5′-tail strand release in the presence of the displaced-strand trap that contains 1 (‘1-Int mismatch Trap’) or two mismatches in the middle of the 40-bp region (‘2-Int mismatch Trap’). The initial rate of unwinding with one mismatched trap is 0.04 ± 0.001 strand/min and with two mismatched trap is 0.02 ± 0.002 strand/min.