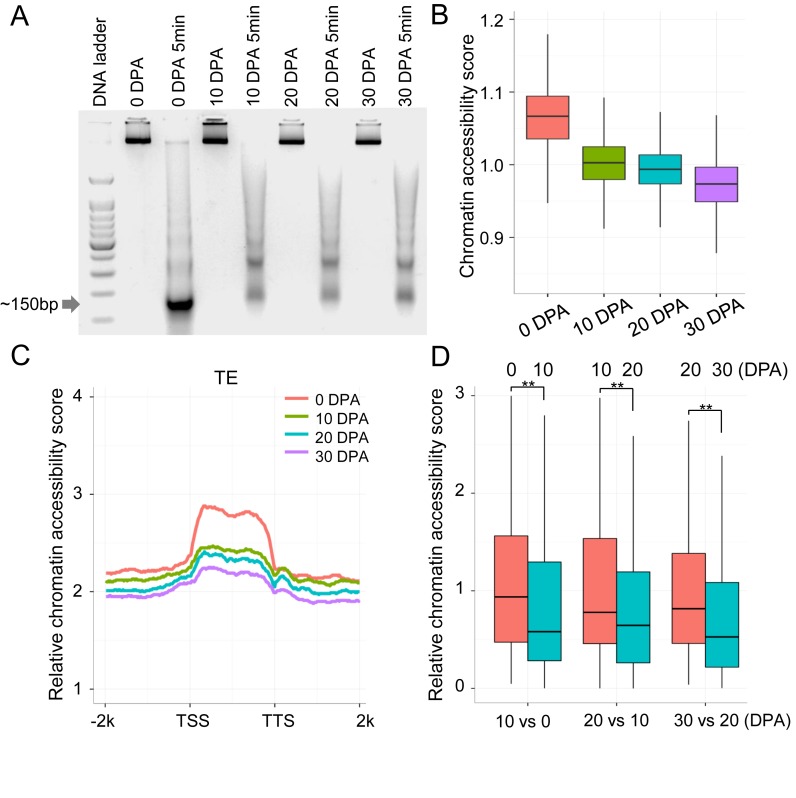
Figure 2.
Chromatin accessibility revealed by MNase-Seq. (A) MNase digestion of chromatin and single nucleosome-bound DNA fragments (∼150 bp) were excised for sequencing. (B) The relative chromatin accessibility in pericentromeric regions of chromosomes. Each pericentromeric region was divided into 100 kb windows. The number of MNase-Seq reads for each window was compared with genome coverage of the control sample. (C) Relative chromatin accessibility of long TEs (>4 kb). (D) Relative chromatin accessibility of hypermethylated regions in CHH context between fibres at 10 DPA and ovules at 0 DPA, between fibres at 20 DPA and fibres at 10 DPA, and between fibres at 30 DPA and fibres at 20 DPA (**P-value < 0.001).