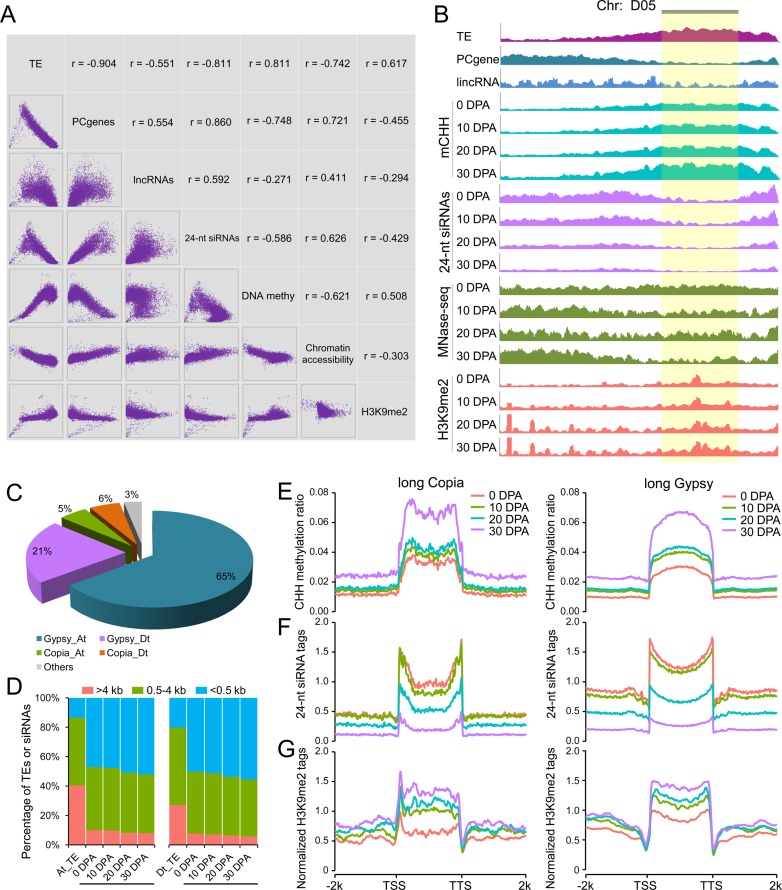
Figure 3.
H3K9me2-dependent pathway contributes to CHH hypermethylation in long TEs. (A) Correlation matrix between H3K9me2 modification patterns with TE content, number of protein-coding genes (PCgenes), number of lincRNAs, number of 24-nt siRNAs, DNA methylation level and chromatin accessibility. Each type of data was measured in 1 Mb windows sliding 200 kb. x and y axes of scatter plots under the diagonal represent the normalized data (log2) in each window. The corresponding pearson correlation for each comparison is indicated above the diagonal. (B) Chromosome landscape (D05) showing several genomic features and H3K9me2 modification patterns. Each type of data was normalized to the same level for comparison between consecutive stages. The pericentromeric region is shown with a gold background. (C) Proportions of Copia and Gypsy long terminal repeat (LTR) retrotransposons occupying long TEs (>4 kb) in the At and Dt subgenomes. (D) Distribution of uniquely mapped 24-nt siRNAs in long TEs (>4 kb, red color), middle-sized TEs (0.5–4 kb, green color) and short TEs (<0.5 kb, blue color). (E) Patterns of CHH methylation level in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb. (F) Patterns of 24-nt siRNA enrichment in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb. (G) Patterns of H3K9me2 ChIP-Seq modification marks in long Copia and long Gypsy TEs from upstream 2 kb to downstream 2 kb.