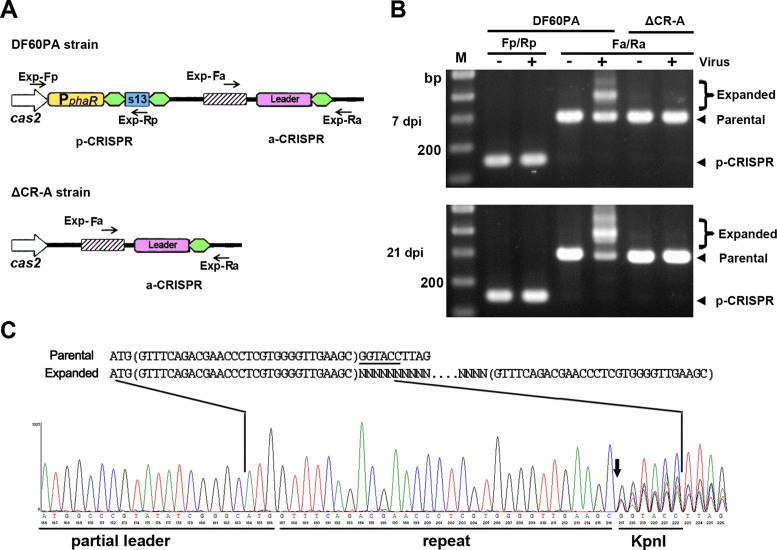
Figure 2.
The leader preceding a single repeat provides sufficient cis-elements for the primed adaptation process. (A) Depiction of the CRISPRs in DF60PA and ΔCR-A. The a-CRISPR carries the 105-bp leader and a single repeat. Exp-Fp/Rp and Exp-Fa/Ra primer pairs (black arrows) were designed against sequences that surround p-CRISPR and a-CRISPR, respectively. (B) PCR assay detecting the expansion of p-CRISPR and a-CRISPR using their corresponding primers. Total DNA from virus-infected (+) or uninfected (-) H. hispanica cells was separately used as the PCR template. For the infected samples, cells were collected 7 or 21 days post infection (dpi). The parental and the expanded a-CRISPRs, respectively, gave rise to ∼300-bp and larger-sized PCR products. Lane Ms, dsDNA size markers. (C) Chromatograph map that shows the sequencing result of the expanded DNA mixture in panel B. The Exp-Fa primer (depicted in panel A) was used for DNA sequencing. The proposed spacer-integration process, in which new spacers are inserted between duplicated repeats (in brackets), is depicted above the chromatograph. Overlap of multiple signals (resulting from the insertion of various new spacers) was observed for positions downstream of the vertical arrow.