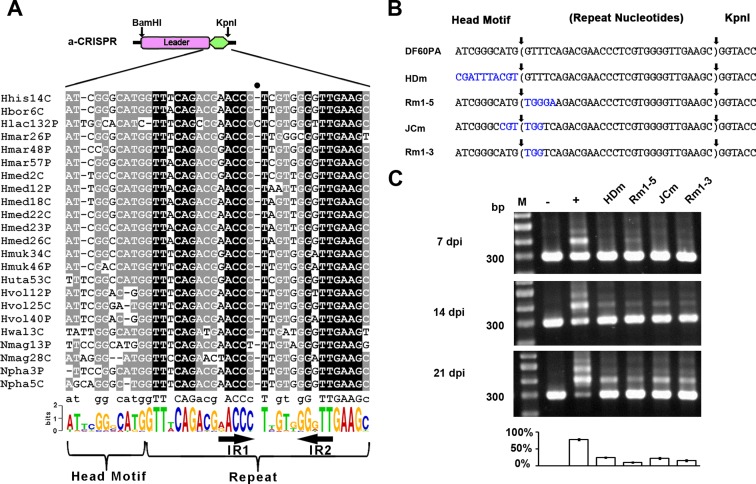
Figure 3.
Sequences spanning the leader-repeat junction are important for adaptation. (A) Sequences spanning the leader-repeat junction are conserved among haloarchaeal CRISPRs with similar repeats. Note that, for each CRISPR, the first repeat and its upstream ∼110-bp sequence were together retrieved and aligned (see Supplementary Figure S2 for details), and only a part of this multi-alignment is shown here. Using the WebLogo server (http://weblogo.berkeley.edu/logo.cgi), a sequence logo was generated from this multi-alignment. The inverted repeats (IR1 and IR2) within the CRISPR repeat are indicated. The full circle (•) indicates the position where an extra nucleotide from the Hlac132P repeat resulted in a gap in all the other repeat sequences during alignment. (B) Illustration of a series of a-CRISPR constructs with mutations (in blue) preceding, following, or flanking the leader-repeat junction. The repeat sequence is shown in brackets. Sequences between vertical arrows were duplicated during spacer integration. (C) Expansion of the a-CRISPRs shown in panel B at 7, 14 or 21 days post HHPV-2 infection (dpi). DNA from the infected (+) or uninfected (–) DF60PA cells was used as the positive or negative control. Three replicates were tested for each mutant, and each gel shows a representative result. The ∼300-bp (parental) and larger-sized (expanded) bands were relatively quantified for the 21-dpi (days post infection) samples, and the percentage of expanded PCR products in each lane is shown in the histogram. Lane Ms, dsDNA size markers.