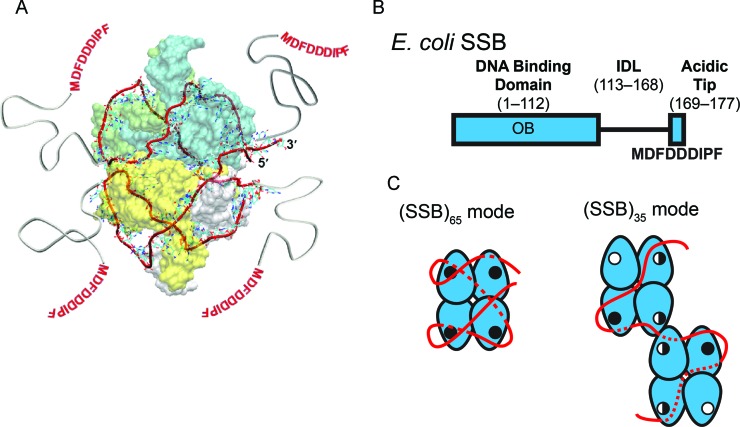
Figure 1.
Structure and binding modes of EcSSB (A) Structural model of 65 nucleotides of ssDNA (red ribbon), wrapped around the EcSSB tetramer (8) in the (SSB)65 mode. (B) Domain organization of EcSSB, depicting the DNA binding domain (OB), the C-terminal IDL and the 9-residue acidic ‘tip.’ (C) The proposed ssDNA binding pathways of the (SSB)65 and (SSB)35 modes of EcSSB (8,12). The EcSSB tetramer is depicted in blue with the ssDNA in red. ssDNA that passes along the backside of the schematic is depicted as a dotted line. OB-fold binding sites are represented with either an open circle, for an unoccupied binding site; a half-closed circled, for a partially occupied binding site; or a closed circle, for a fully occupied binding site (see Discussion for details of this model). Two EcSSB tetramers are shown in the (SSB)35 mode to denote high cooperativity. The C-terminal tails are not depicted for clarity.