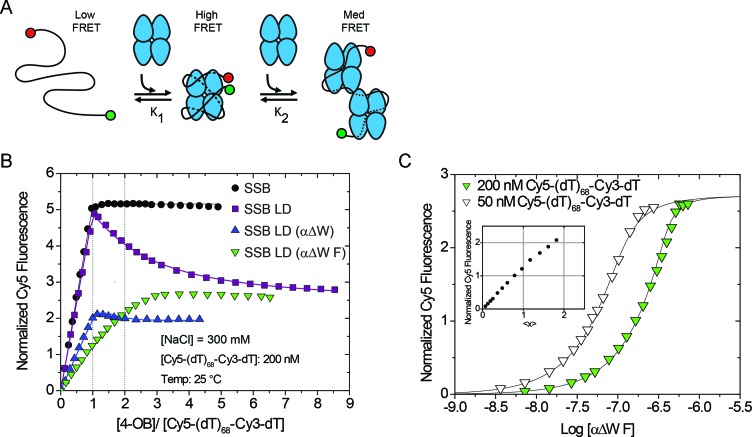
Figure 4.
Mutants with only two wild-type-like OB folds do not form a fully wrapped complex with ssDNA. (A) Reaction scheme of SSB constructs titrated into Cy5-(dT)68-Cy3-dT, a substrate that is sensitive to the degree of wrapping and binding density of SSB:ssDNA complexes. (B) Results of equilibrium titrations of protein titrated into 200 nM Cy5-(dT)68-Cy3-dT in Buffer T + 300 mM NaCl at 25°C plotted as normalized Cy5 fluorescence versus the ratio of total protein to DNA concentrations. Under these conditions, wtSSB binds tightly with Cy5-(dT)68-Cy3-dT (Kobs ≥ 1010 M−1) in a one-to-one complex with a maximum normalized enhancement of 5.16 ± 0.01. The proteins SSB LD, and SSB LD (αΔW) first bind tightly to Cy5-(dT)68-Cy3-dT in a one-to-one complex (Kobs ≥ 5 × 1010 M−1) then a second SSB tetramer binds more weakly [(3.8 ± 0.3) × 106 M−1 and (2 ± 2) × 108 M−1, respectively]. Under these conditions, the SSB LD (αΔW F) construct does not clearly transition through a simple one-to-one complex. (C) To measure the binding of SSB LD (αΔW F) more rigorously, we also performed the titration into 50 nM 5′-Cy5-(dT)68-Cy3-dT-3′ and globally fit the data to a two-site binding model. The first molecule of SSB LD (αΔW F) binds with Kobs = (1.4 ± 0.7) × 108 M−1 with a normalized Cy5 fluorescence of 1.18 ± 0.06 and the second molecule binds with Kobs = (4 ± 1) × 107 M−1 with a normalized fluorescence of 2.71 ± 0.06. Inset shows the results of a model independent binding analysis which plots the normalized Cy5 fluorescence against the number of SSB LD (αΔW F) tetramers bound per ssDNA (<x>).