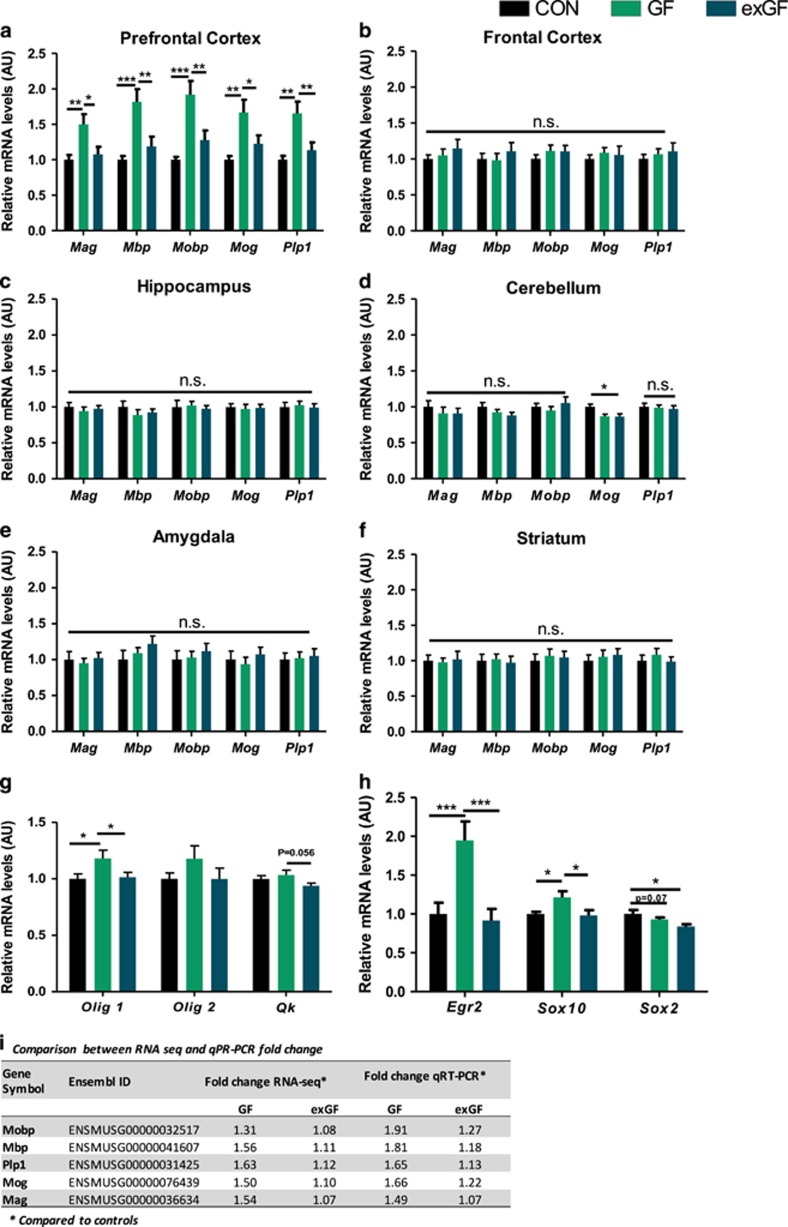
Figure 2.
Quantitative real-time PCR (qRT-PCR) validations of RNA-seq data within various brain regions of selected myelin component genes were found to be brain region specific. Germ-free status in mice results in increased myelin gene expression only in the PFC, which was normalized in exGF mice. (a–f) qRT-PCR of myelin gene transcripts and regulatory factors in the prefrontal cortex, frontal cortex, hippocampus, cerebellum, amygdala and striatum. Bar graphs indicate average values in 12 mice per group after β-actin normalization relative to average control levels. (g) Significant changes in oligodendrocyte-specific genes. (h) Changes in known genes involved in regulation of myelination. (i) Table representing RNA-seq and qRT-PCR fold change for individual myelin component genes used for RNA-seq validation. Fold change is in comparison with the control group. (a, g, h) Prefrontal cortex, (b) frontal cortex, (c) hippocampus, (d) cerebellum, (e) amygdala and (f) striatum. Data graphed as ±s.e.m. *P<0.05; **P<0.01; ***P<0.001.