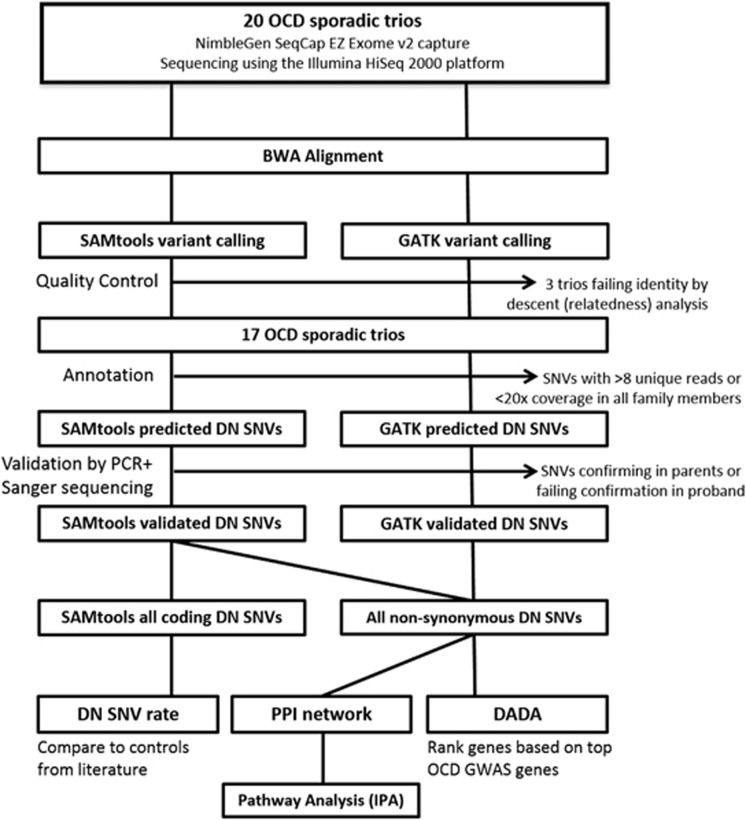
Figure 1.
Single-nucleotide variant (SNV) discovery, quality control, annotation and analysis workflow. Whole-blood samples from obsessive-compulsive disorder (OCD) probands and their unaffected parents were enriched for exonic sequence with the NimbleGen SeqCap EZ Exome capture reagents and sequenced using the Illumina HiSeq 2000 platform. Identity by descent analysis was performed to confirm relatedness among samples. Final analyses included 17 OCD trios. Only de novo (DN) SNVs called by SAMtools and validated by Sanger sequencing (present in proband and absent in parents) were carried into DN SNV rate analyses. For subsequent analyses of protein–protein interaction (PPI), Degree-Aware Disease Gene Prioritization (DADA) and Ingenuity Pathway Analyses (IPA), we also included confirmed DN SNVs from a second alignment and variant calling pipeline, which followed the GATK v3 best practices guidelines.