Figure 6.
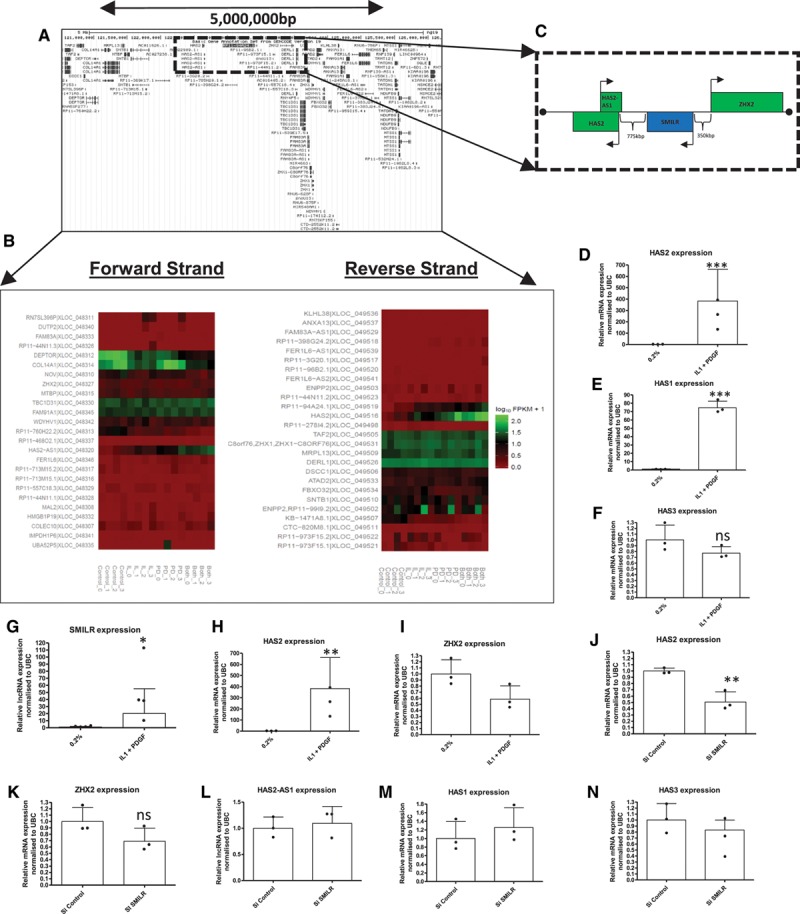
SMILR regulates proximal gene HAS2 in chromosome 8. A, Schematic view of the 8q24.1 region showing lncRNAs and protein-coding genes over the 5 000 000-bp region from Ensemble. B, Regulation of protein-coding and noncoding genes within the selected region following IL1α and PDGF treatment; heatmap depicts expression of genes found with RNA-seq in 4 patient samples. C, Dotted line marks region containing SMILR lincRNA and closest protein-coding genes HAS2 and ZHX2. D, Expression of proximal gene HAS2 – modulated in the same manner as SMILR with IL1α and PDGF treatment (n=3). Unpaired t test: ***P<0.001 vs 0.2%. E and F, Additional HAS isoforms are differentially modulated by IL1 and PDGF (n=3). Unpaired t test: ***P<0.001 vs 0.2%. G through I, Validation of RNA-seq data for lncRNAs SMILR and HAS2-AS1 by qRT-PCR (n=3). *P<0.05 and **P<0.01 vs 0.2%, unpaired t test. J, Inhibition of SMILR expression via dsiRNA treatment significantly inhibits HAS2 expression determined by qRT-PCR **P<0.01 vs Si control. Unpaired t test (n=3). K through N, SMILR inhibition had no effect on proximal genes ZHX2 or HAS2-AS1 nor additional HAS isoforms, HAS1 or HAS3 (n=3). Unpaired t test. ANOVA indicates analysis of variance; dsiRNA, dicer substrate small interfering RNA; HSVSMC, human saphenous vein–derived smooth muscle cell; IL1α, interleukin-1α; lincRNA, intervening long noncoding RNA; lncRNA, long noncoding RNA; PDGF, platelet-derived growth factor; qRT-PCR, quantitative real-time polymerase chain reaction; Si, small interfering; and UBC, ubiquitin.
