Fig 3. Maximum likelihood phylogenetic tree of full-length cotton leaf curl geminiviruses: The phylogenetic tree was generated for cotton-infecting begomoviruses.
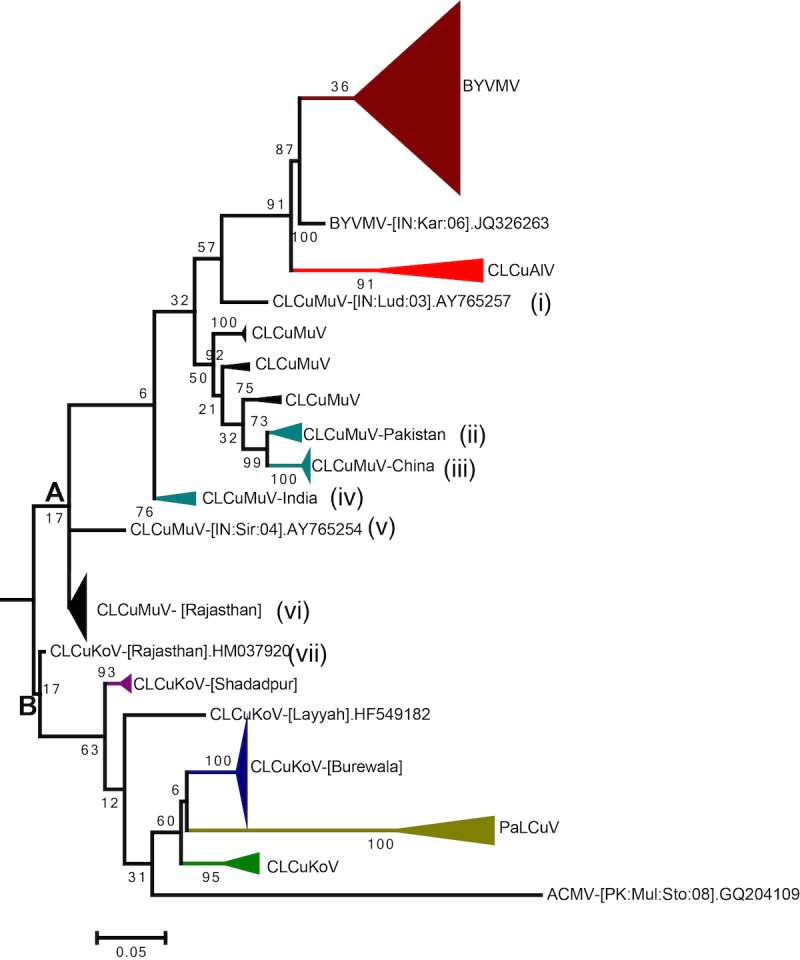
Group-A represents the major cotton-infecting viruses whose sequences are mainly derived from CLCuMuV. Group-B represents viruses whose sequences are mainly derived from CLCuKoV. Seven different clades are found because of recombination between CLCuMuV and CLCuKoV, or BYVMV. Each clade shows a specific recombination pattern (see Fig 2). African cassava mosaic virus was chosen as an out-group in the phylogenetic analysis. The numbers at the nodes represent bootstrap values.
