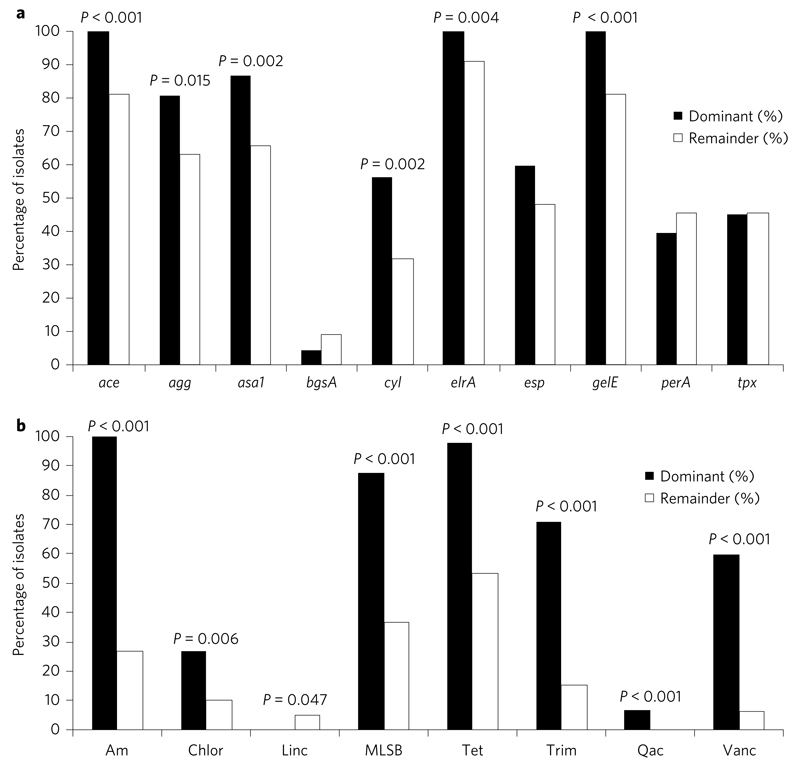
Figure 3. Prevalence of virulence and antibiotic resistance genes in the dominant lineages (L1–L3, n = 89) and remainder (n = 79).
a,b, Graphs showing percentage of isolates for which putative virulence genes (a) or antibiotic resistance genes (grouped by antibiotic class) (b) were detected. Genes that were ubiquitous in the collection are not shown. P values are shown when a significant difference was observed using Fisher’s exact test. Virulence genes: ace = collagen adhesion protein; agg = aggregation substance; asa1 = aggregation substance; bgsA = biofilm-associated glycolipid synthesis A; cyl = cytolysin; elrA = enterococcal leucine-rich protein A; esp = enterococcal surface protein; gelE = gelatinase; perA = pathogenicity island-encoded regulator; tpx = thiol peroxidase. Antibiotic resistance genes: Am = aminoglycosides (comprising one or more of aac6′-2″, aph3″-III, aacA, ant-6-Ia, str); Chlor = chloramphenicol (cat); Linc = lincosamides (lnuB); MLSB = macrolide, lincosamide, streptogramin B (comprising ermB or ermT); Tet = tetracycline (comprising one or more of tetL, tetM, tetO, tetS); Trim = trimethoprim (comprising dfrC, dfrD, dfrF or dfrG); Qac = quaternary ammonium compounds and other antiseptics (qacZ); Vanc = vancomycin.