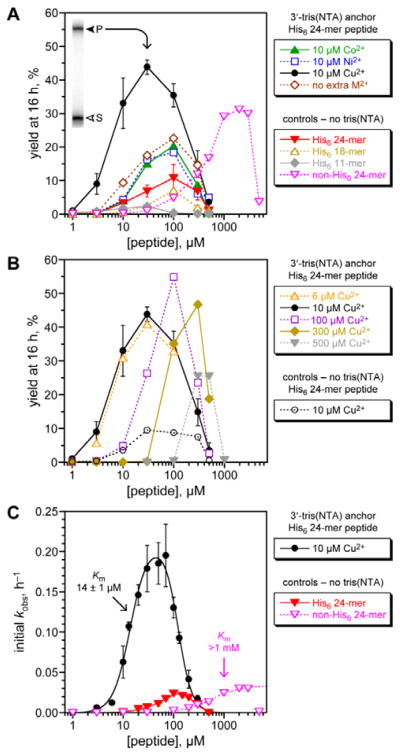
Fig. 3.
Assessing histidine tag recruiting for the 8XJ105 deoxyribozyme. See the Experimental section for explanation of error bars in all panels. Conditions: 20 nM 3′-32P-radiolabeled 5′-pppRNA, 0.5 μM 8XJ105 deoxyribozyme, 2 μM 3′-tris(NTA) DNA anchor oligonucleotide, 1–5000 μM peptide, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 1 mM ZnCl2, 150 mM NaCl, and CoCl2, NiCl2, or Cu(NO3)2 as indicated at room temperature. CuCl2 gave equivalent outcome. (A) Yield at 16 h of peptide-RNA conjugate as a function of concentration of His6-tagged 24-mer peptide (or untagged 24-mer in one case). All experiments including the controls were performed with 1 mM Zn2+; 10 μM Co2+, Ni2+, or Cu2+ was additionally present where indicated. Inset: PAGE data at 16 h for 3′-32P-radiolabeled RNA and 30 μM His6-tagged 24-mer peptide with Cu2+ (S = substrate, P = product). See Fig. S1 (ESI†) for data with 5′-tris(NTA) moiety. 24-mer H6SAGERASAEDMARAAYAA (for non-His6, replace H6 with ASAASA); 18-mer H6SAEDMARAAYAA; 11-mer H6AAYAA. (B) Optimization of Cu2+ concentration, and demonstration that Cu2+ alone [without tris(NTA)] is not responsible for the recruiting effect. (C) Determination of peptide Km values using initial-rate kinetics. See curve fit descriptions and fit details in the Experimental section.