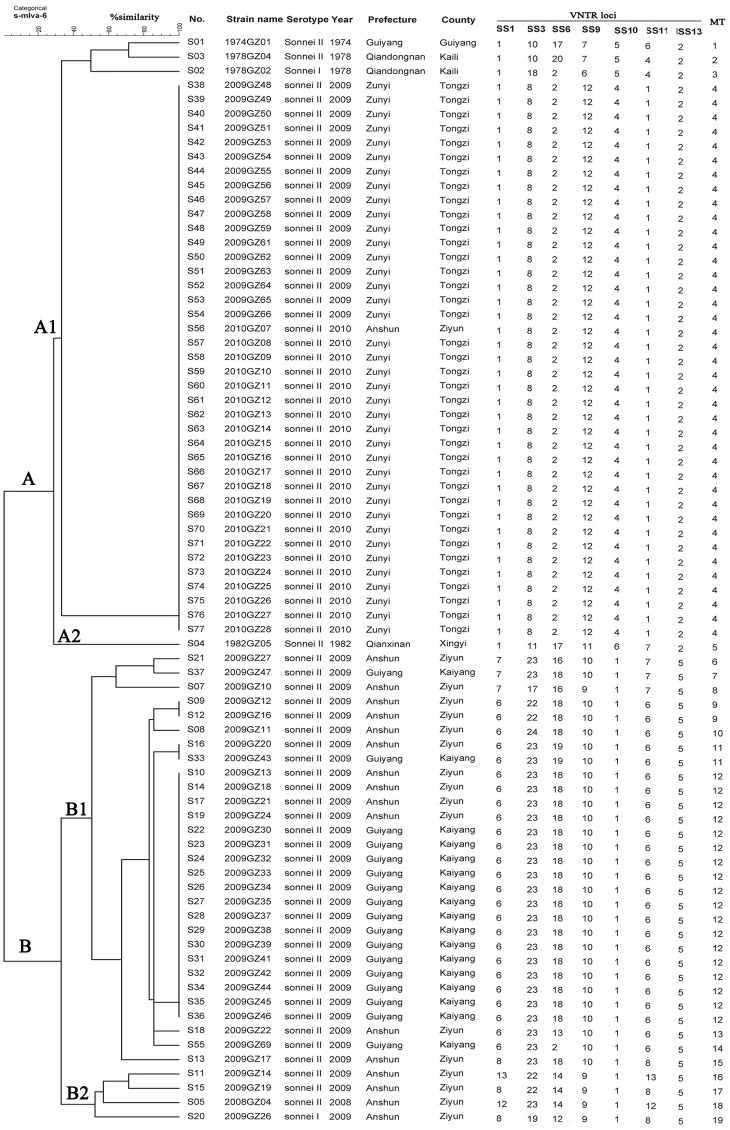
Fig 2. Relationships of 76 Shigella sonnei isolates from Guizhou Province based on the Multiple-Locus Variable number tandem repeat (VNTR) Analysis (MLVA).
The 76 S. sonnei isolates from Guizhou Province were analyzed by MLVA based on seven VNTR loci. The copy number of the repeats of each VNTR locus was deduced from the length of the amplicons. This number was then converted into an allele designation, which in turn formed the allele string for the seven loci. The allele string was constructed in the following order: SS1-SS3-SS6-SS9-SS10-SS11-SS13. The data were incorporated into BioNumerics software and analyzed. Each unique allelic string was designated a unique MLVA type (MT). A dendrogram constructed using the PFGE patterns was generated by the UPGMA algorithm using the Dice-predicted similarity value of two patterns. The corresponding MLVA type with copy numbers for the seven VNTRs, serotype, and background information were shown alongside the dendrogram on the right.