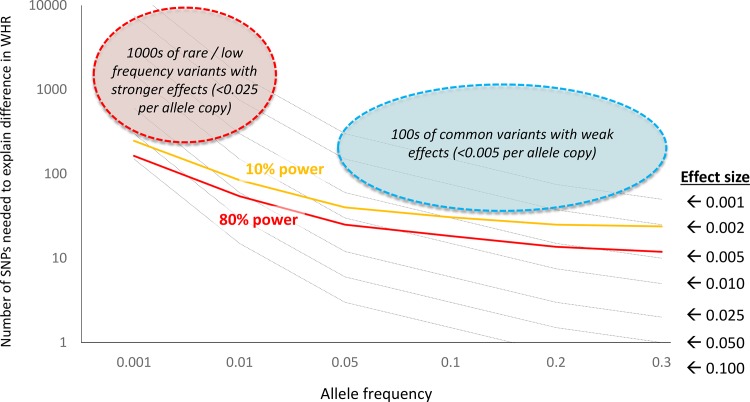
Fig 2. Contribution of population-specific genetic variants to increased WHR amongst South Asians compared to Europeans.
Results are shown as the number of SNPs needed to fully explain the difference in WHR between the populations across a range of effect sizes (per allele copy, dashed black lines). Superimposed are lines showing the power (10% power, orange line; 80% power, red line) of the current study to detect common and low frequency genetic variants associated with WHR amongst South Asians. Results show that our study sample size is sufficient to identify common variants with effect size >0.005 per allele copy, and rare / infrequent variants with effect size >0.025 (untransformed β values). At these effect sizes, there would need to be 10s to 100s of population-specific genetic variants to explain increased WHR amongst South Asians. At effect sizes smaller than those identifiable in the current study, there would need to be 100s of common variants or 1000s of rare / low frequency variants that are population-specific and associated with WHR, to account for increased WHR amongst South Asians.