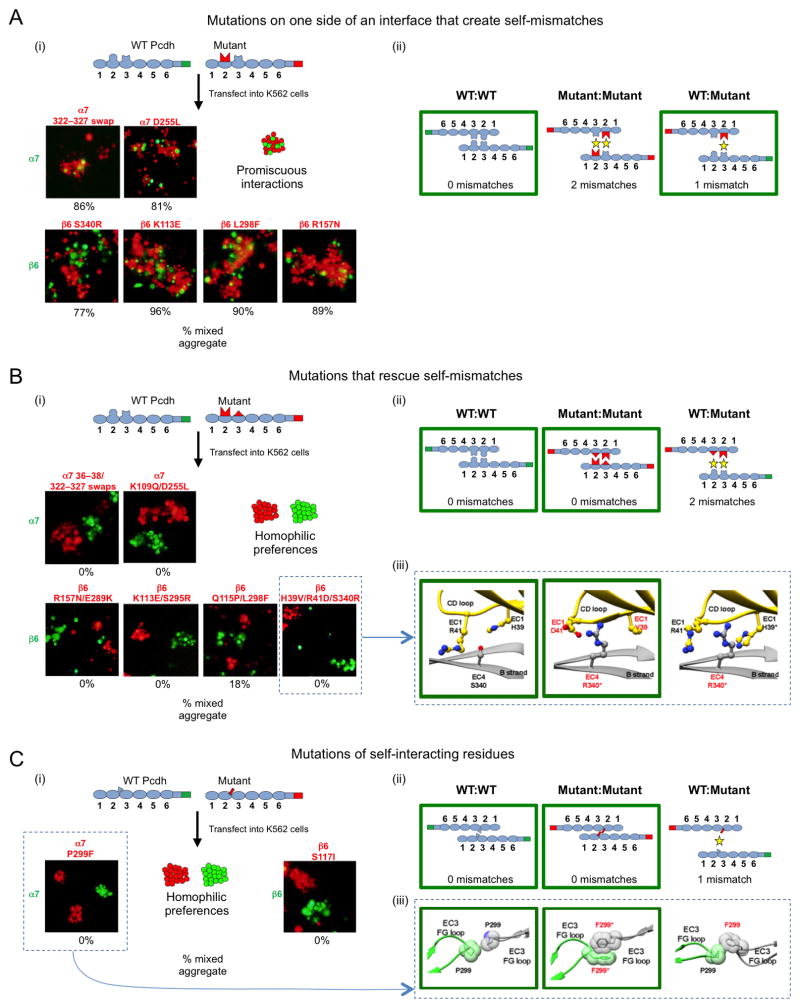
Figure 5. Pcdh homophilic recognition depends on trans-interacting variable residues.
A. Single-sided α7 or β6 interface mutants were assayed for binding specificity in a mixed cell aggregation assay with wild-type (WT) α7 or β6. (i) Schematic of WT mVenus-tagged Pcdh and mutant mCherry-tagged Pcdh (top). Representative images of mixed cell aggregation assays are shown for each mutant (bottom). In all cases mixed aggregates were observed. The percentage of mixed (red/green) aggregates, from three independent experiments, is given below each image. (ii) Schematic illustrations of the three possible Pcdh trans dimers that can form between the interacting cells. In each case, the number of expected mismatched residues is indicated. The green boxes highlight the interactions that mediate the observed aggregates in panel (i).
B. Complementary (two-sided) mutations of trans-interacting residues were introduced in α7 or β6, and binding specificity was assessed in the mixed cell aggregation assay. (i) Separate aggregates of WT and mutant Pcdh expressing cells were formed in all cases. (ii) The schematic shows that the putative mutant:WT interaction would be expected to contain two mismatched residue interactions, explaining why mixed aggregates were not observed. (iii) Structural models for the two homophilic (WT:WT and mutant:mutant) interactions that mediate the observed cell aggregates, and the putative heterophilic (WT:mutant) interface, which does not form, are shown for the β6 WT with β6 H39V/R41D/S340R mutant case.
C. Mutations of residues that interact symmetrically between the two protomers. (i) Separate aggregates of WT and mutant cells were formed in all cases indicating that a new homophilic Pcdh has been generated. (ii) As in (B), homophilic preferences of WT and mutant proteins can be explained by the presence of mismatched residue interactions in the WT:mutant complex. (iii) Structural models are shown for the potential α7 WT and α7 P299F mutant complexes.
See also Figure S5.