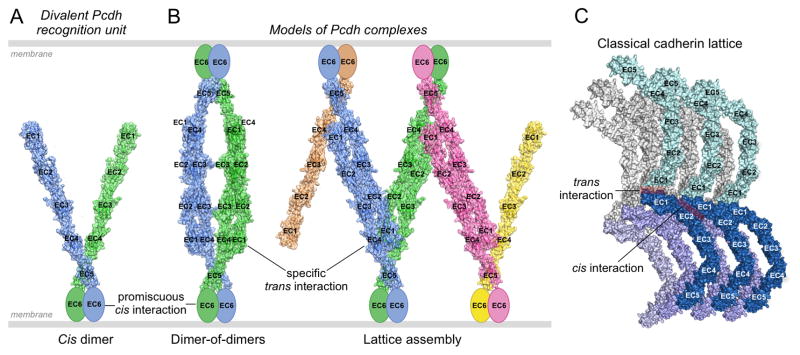
Figure 6. Pcdh-mediated recognition units and possible binding modes between opposed cell surfaces.
A. Model of a cis-dimeric Pcdh recognition unit. The dimerization interface is located on EC6 which is depicted as an ellipse since no structure for this domain is available. Each of the two arms is taken from the 5-domain structure of α7EC1–5. The angle between them is arbitrary. The two arms are depicted in different colors since they may correspond to different isoforms.
B. Models of possible Pcdh recognition complexes. The left panel depicts a dimer of recognition dimers yielding a tetrameric complex. Although the two arms are depicted here as different, the only tetramer detected so far (in solution, Rubinstein et al., 2015) has both arms identical. The right panel depicts the initiation of a linear zipper formed from different recognition units on each membrane that contain one common isoform. Mismatched isoforms, expressed by one cell but not the other, are represented in pink and yellow, and in principle would terminate growth of the intercellular Pcdh zipper.
C. Classical cadherin intercellular assembly, corresponding to the extracellular structure of adherens junctions, mediated by cis and trans interactions.