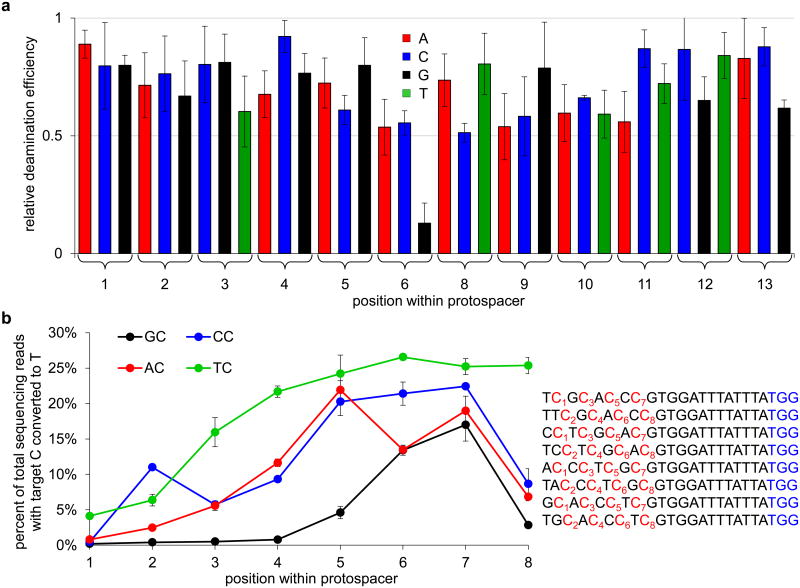
Figure 2. Effects of sequence context and target C position on base editing efficiency in vitro.
a, Effect of changing the sequence surrounding the target C on editing efficiency in vitro. The deamination yield of 80% of targeted strands (40% of total sequencing reads from both strands) for C7 in the protospacer sequence 5′-TTATTTCGTGGATTTATTTA-3′ was defined as 1.0, and the relative deamination efficiencies of substrates containing all possible single-base mutations at positions 1-6 and 8-13 are shown. b, Positional effect of each NC motif on editing efficiency in vitro. Each NC target motif was varied from positions 1 to 8 within the protospacer as indicated in the sequences shown on the right. The PAM is shown in blue. The graph shows the percentage of total DNA sequencing reads containing T at each of the numbered target C positions following incubation with BE1. Note that the maximum possible deamination yield in vitro is 50% of total sequencing reads (100% of targeted strands). Values and error bars reflect the mean and standard deviation of three (for a) or two (for b) independent biological replicates performed on different days.