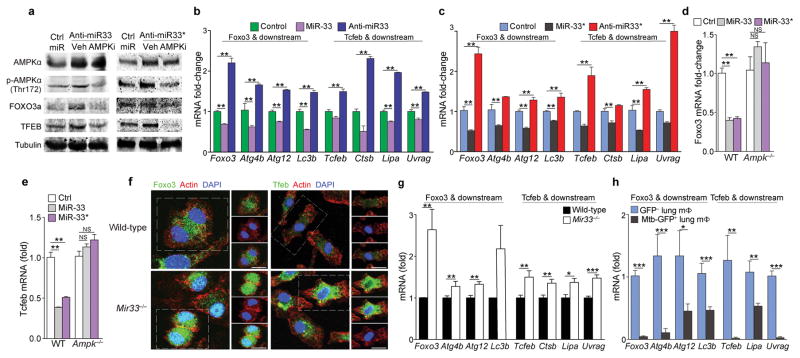
Figure 4. miR-33 and miR-33* repress AMPKα and downstream transcription factors controlling autophagy and lysosomal gene programs.
(a) Western blotting of lysates from peritoneal macrophages treated with anti-miR-33 or anti-miR-33* or control (ctrl) miR in the presence or absence of the AMPK inhibitor compound C (Ampki, 5 μM). (b, c) mRNA levels of Foxo3, Tcfeb and their downstream transcriptional targets in peritoneal macrophages transfected with (b) miR-33 and (c) miR-33* mimics or inhibitors as indicated. (d) Foxo3 and (e) Tcfeb mRNA in wild-type (WT) or Ampk−/− BMDMs treated with mmu-miR-33, -33*, or Ctrl mimic. (f) Immunofluorescence (IF) imaging of FOXO3a and TFEB in green, F-actin (red) and DAPI (blue) in wild-type (WT) and Mir33−/− BMDMs. (g) Quantification of Foxo3, Tcfeb and their downstream transcriptional targets (right) in WT and Mir33−/− BMDMs. (h) Quantification of Foxo3, Tcfeb and their downstream transcriptional targets in GFP+ (infected) or GFP− (uninfected) alveolar macrophages isolated from mice infected with a GFP-expressing H37Rv Mtb strain, 2 weeks post-infection. *P≤0.1, **P≤0.05, ***P≤0.005 (Student’s t-test (b–c, g, h), one-way ANOVA (d, e)). Data are from one experiment representative of 3 ((b, c, g; mean ± s.e.m)) or 2 ((a, f) independent experiments. Data are from 2 experiments (d, e; mean ± s.e.m). Data are from 3 mice (h; mean ± s.e.m).