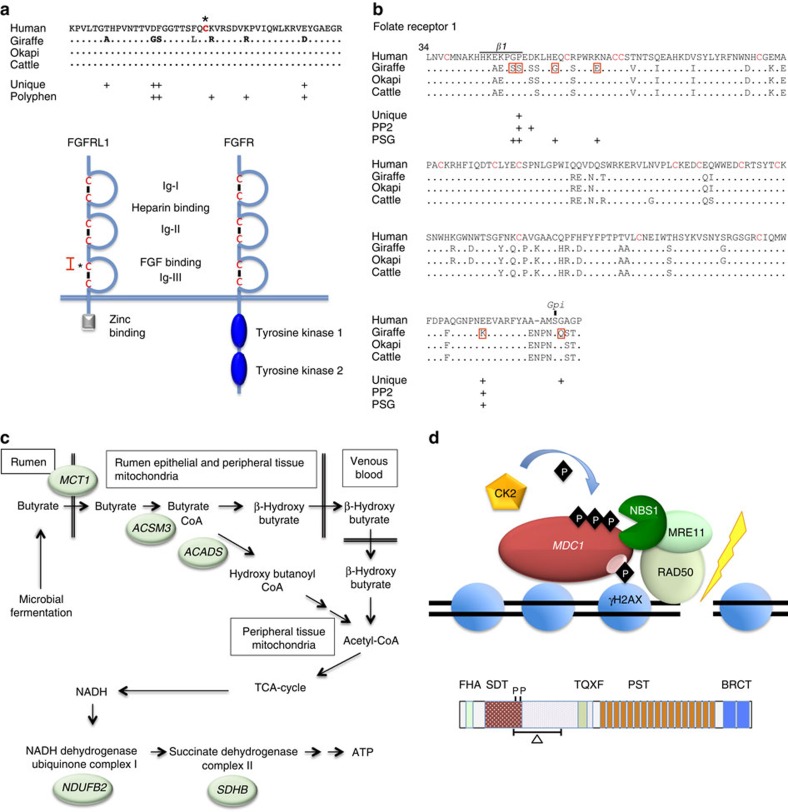
Figure 3. Giraffe genes and pathways exhibiting extraordinary divergence and patterns of amino acid substitutions.
(a) Giraffe FGFRL1 contains seven amino acid substitutions that are unique at fixed sites in other mammals and/or are predicted by Polphen2 analysis to alter function (upper panel). Human reference is shown, which is identical to cattle and okapi in this segment. The unique giraffe substitutions occur in the FGF-binding domain region flanking the N-terminal cysteine (asterisk) of the Ig-III loop (lower panel). Red bracket in lower panel corresponds to the sequence in the upper panel. The extracellular structure of FGFRL1 (left) is the same as a prototypical FGF receptor (FGFR, right) but lacks the cytoplasmic C-terminal tyrosine kinase domains seen in FGFR and instead contains a zinc-binding domain. (b) Giraffe FOLR1 contains seven substitutions that each show evidence of positive selection (P<0.05) by the branch-site model. Two of the positive selected sites (PSG), P48S and E222K, are also unique substitutions at fixed sites and Polyphen2 (PP2) analysis predicts them to alter function. P48S is within β-sheet-1 that forms part of the folic acid-binding pocket. The FOLR1 protein forms a globular structure maintained by overlapping disulfide bridges between 16 cysteine residues (red) and tethered to the plasma membrane at S233 by a Gpi anchor. The unique substitution in giraffe, G234Q, immediately adjacent to the Gpi anchor site may alter the anchor site or the rate of its formation. (c) Genes encoding key enzymes in butyrate metabolism and downstream mitochondrial oxidative phosphorylation pathways have diverged in giraffe including the monocarboxylate transporter (MCT1), acyl-coenzyme A synthetase-3 (ACSM3), short-chain specific acyl-CoA dehydrogenase (ACADS), NADH dehydrogenase (ubiquinone) 1β subcomplex subunit 2 (NDUFB2) and succinate dehydrogenase [ubiquinone] iron-sulfur subunit (SDHB). ACSM3 and ACADS are located in the mitochondrial matrix where as NDUFA2, NDUFB2 and SDHB are located in the mitochondrial inner membrane. In addition to being present in the rumen epithelial cells, MCT1 is highly expressed in the heart, skeletal muscle and the nervous system where it acts to transport volatile fatty acids (VFAs) and lactate. (d) Double-strand break repair genes exhibit divergence in giraffe and/or okapi. The mediator of DNA-damage check point 1 (MDC1) binds phosphorylated H2AX, which mark DNA double-strand break, and serves as scaffold to recruit the MRN DNA repair complex composed of NBS1, MRE11 and RAD50 (upper panel). The giraffe and okapi MDC1 gene exhibits a 264 amino acid deletion that removes part of the SDT region that harbours two critical CK2 phosphorylation sites (lower panel). These two phosphorylation sites are among multiple sites that regulate the interaction of MDC1 and NBS1 essential for the recruitment of the MRN complex to double-strand breaks.