Abstract
BRCA1-associated RING domain protein 1 (BARD1) stabilizes BRCA1 protein by forming a heterodimeric RING-RING complex, and impacts function of BRCA1, including homologous recombination (HR) repair. Although colon cancer cells usually express wild type BRCA1, presence of an oncogenic BARD1 splice variant (SV) in select cancers may render BRCA1 dysfunctional and allow cells to become sensitive to HR targeting therapies. We previously reported association of loss of full-length (FL) BARD1 with poor prognosis in colon cancer as well as expression of various BARD1 SVs with unknown function. Here we show that loss of BARD1 function through the expression of a BARD1 SV, BARD1β, results in a more malignant phenotype with decreased RAD51 foci formation, reduced BRCA1 E3 ubiquitin ligase activity, and decreased nuclear BRCA1 protein localization. BARD1β sensitizes colon cancer cells to poly ADP ribose polymerase 1 (PARP-1) inhibition even in a FL BRCA1 background. These results suggest that expression of BARD1β may serve as a future biomarker to assess suitability of colon cancers for HR targeting with PARP-1 inhibitors in treatment of advanced colon cancer.
Colorectal cancer (CRC) is the third most common cancer worldwide, and in 2014, there were an estimated 50,710 deaths from CRC in the U.S.1,2. Despite improvements in early detection, over half of the cases are diagnosed at advanced stages which are associated with a poor outcome. Recent data show a trend towards an increase in metastatic cases in young patients, underscoring the need for targeted therapeutic options in advanced disease3. Examination of a large cohort of colon carcinomas showed that homologous recombination (HR) repair is deficient in greater than 78% of tumors4. Therefore, therapeutics which target HR deficiencies may be effective in colon cancer.
BRCA1 is a tumor suppressor which displays E3 ubiquitin ligase activity and has been associated with HR repair, cell cycle checkpoint regulation, and chromatin dynamics5,6. BRCA1 is stabilized by forming a heterodimer with BRCA1-associated RING domain protein 1 (BARD1) which binds through their RING finger domains7. Dimerization not only stabilizes the two monomers, but is also essential for BRCA1’s tumor suppressive activities8,9. Therefore, mutations affecting the BARD1/BRCA1 heterodimer structure may confer an increase in cancer risk. Notably, germline mutations in BRCA1 are associated with a predisposition to breast and ovarian cancers10,11 and, while rare, germline mutations in BARD1 also contribute to hereditary and sporadic breast and ovarian cancer12. BRCA1 is usually wild type in sporadic colon cancers, however, germline mutations to BRCA1 have been described to be increased in early onset colon cancers13.
HR defective cells, such as breast cancer cells harboring mutant BRCA1, display greater than normal sensitivity to poly ADP ribose polymerase 1 (PARP-1) inhibition14. Treatment with PARP-1 inhibitors (PARPi) results in an increase in single-strand DNA break formation15,16,17, which if they persist unrepaired, collision of unresolved single strand breaks with replication forks is thought to result in double-stranded breaks (DSB)18,16. Binding of BARD1-BRCT domain to PAR motifs is important for the early recruitment of BRCA1 to DNA damage sites19 and the BARD1/BRCA1 heterodimer plays an important role in the repair of DSB by HR20. In addition, BARD1/BRCA1 dimers exhibit ubiquitin ligase activity, which also targets proteins of the RNA polymerase complex and they are subsequently degraded by the proteasome to initiate HR repair21. If HR does not function properly and DSBs are not repaired, cell death can occur14.
Typically in colon cancer, BRCA1 is wild type, but we have reported that FL BARD1 expression may be lost, which is associated with poorer outcome. Further, various, differentially expressed BARD1 SVs have been reported7,22,23. In colon cancer, we observed a BARD1 SV, BARD1β which lacks exons 2 and 3, encoding most of the RING finger domain22. BARD1β has also been reported to be expressed in neuroblastoma, non-small cell lung cancer, breast and ovarian cancers21,22,23,24. While FL BARD1 is a tumor suppressor, BARD1β has been reported to be pro-proliferative and tumor initiating22,23,24. Its overexpression leads to neoplastic cell transformation in murine fibroblasts, and its repression leads to growth arrest of cancer cells22,24,25. BARD1β expression has been reported to correlate with poor survival in colon cancer23. However, insights into the precise mechanisms of how isoforms may contribute to tumorigenesis in colon cancer and specifically how knowledge of expression may be harnessed in treatment strategies of colon cancer patients are lacking.
In this study, we demonstrate that the expression of the BARD1β correlates with impaired HR in PARPi-sensitive colon cancer cells. Moreover, exogenous expression of the BARD1β isoform in PARPi-resistant colon cancer cells lead to a more transformed phenotype and impaired HR. These cells also display an increased sensitivity to PARPi, suggesting possible therapeutic applications for cancer patients expressing the BARD1β. Treatment with PARPi in conjunction with the DNA-damaging chemotherapeutic agents, Irinotecan or Oxaliplatin, leads to an increased sensitivity to both agents when BARD1β is expressed. We conclude that BARD1β merits further investigation as a therapeutic biomarker for the use of adjunct PARPi therapy in advanced colon cancer.
Results
Human colon cancer cells exhibit differential sensitivity to PARP-1 inhibition
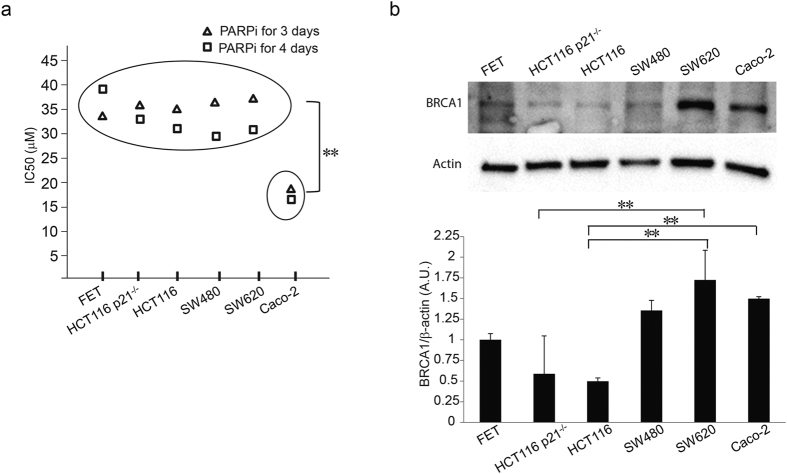
Increased sensitivity to PARP-1 inhibition is indicative of defective DNA double-strand break repair by homologous recombination16,26. To determine etiologies of PARPi sensitivities in colon cancer cells, we treated a panel of colon cancer cell lines representing a spectrum of genomic backgrounds with increasing concentrations of PARPi (Fig. 1a). Cell viability and dose response curve for each cell line was determined after 3 and 4 days of treatment and IC50s were calculated. None of the known genomic determinants of colon carcinogenesis, including microsatellite instability (MS), loss of SMAD4 or loss of p21, were associated with an increased sensitivity to PARPi. Yet the MS stable Caco-2 cell line was significantly more sensitive to PARPi, suggestive of an undefined distinct molecular feature in these cells (Table 1).
Figure 1. Human colon cancer cells exhibit differential sensitivity to PARP-1 inhibition.
(a) CRC cell lines FET, HCT116 p21−/−, HCT-116, SW480, SW620, and Caco-2 were treated with PARPi for 3 (triangle) and 4 (square) days and IC50 determined (**p < 0.01). The experiment was performed at least three independent trials with at least three technical replicates. (b) Untreated CRC cell lines were harvested, protein extracts were prepared, separated and immunoblotted with anti-BRCA1 antibody as described in Methods. The experiment was repeated at least three times. Quantification of protein band intensity was performed by ImageJ software (*p < 0.05 and **p < 0.01).
Table 1. Colon cell lines listed with genetic backgrounds.
| Cell line | Characteristic |
|---|---|
| FET | Early stage CRC, well diff., high TGFβ1 |
| HCT116 p21−/− | p21 negative cell line HCT116 |
| HCT116 | Mismatch repair deficiency |
| SW480 | SMAD4 null |
| SW620 | SW480 lymph node metastatic derivative |
| Caco-2 | p21 and SMAD4 wild type, MS stable |
Differential sensitivity to PARPi among colon cancer cell lines is not associated with BRCA1 mutation or loss of BRCA1 expression
BRCA1 is a major participant in HR, therefore cells deficient in BRCA1 show increased lethality with PARPi due to absence of DSB DNA repair by HR. All tested colon cancer cell lines are known to have wild type BRCA1 and all express FL BRCA1 protein. BRCA1 protein was in similar quantities among SW620, Caco-2, and SW480 cells and these were relatively higher than FET, HCT116 p21−/−, HCT116 (Fig. 1b).
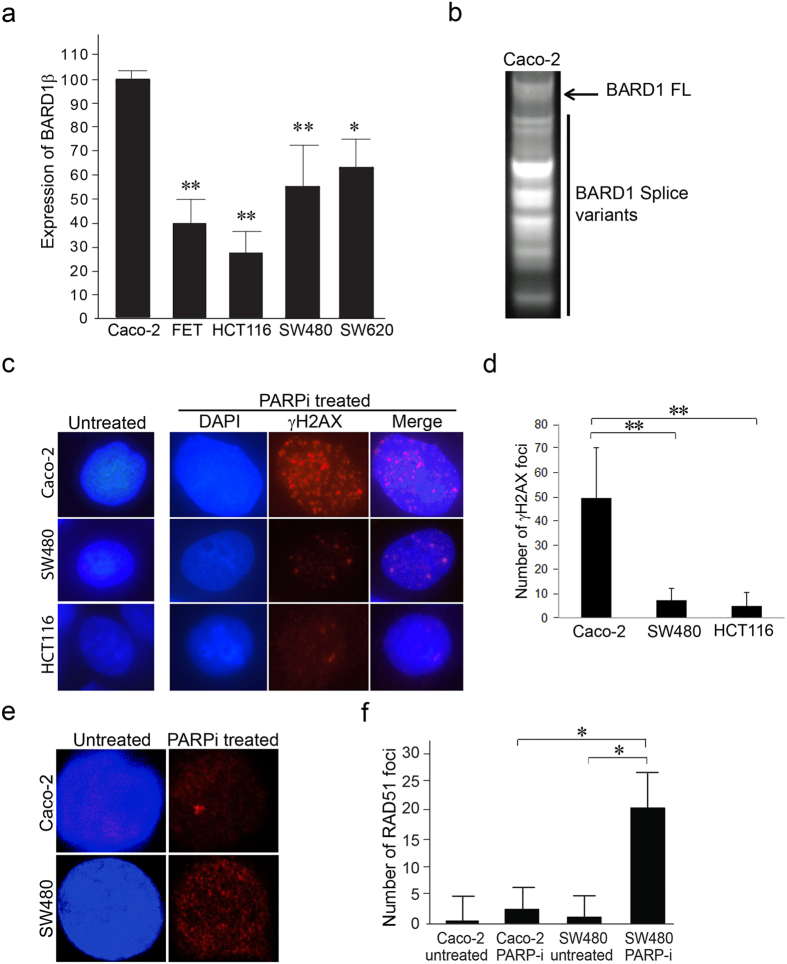
Alterations in BARD1 may affect BRCA1 function in a BRCA1 wild type setting. FL BARD1 is present in both PARPi-sensitive and -resistant cell lines. However, BARD1 SVs may have functions independent from its FL protein and may affect the function of BRCA1 independent of their effects on BARD1 expression. BARD1β has been reported to have poor prognosis in colon cancer22. Using real-time PCR, we compared the expression of BARD1β in a panel of colon cancer cell lines. Expression of BARD1β was significantly higher in PARPi sensitive Caco-2 cells compared to its expression in cell lines that were resistant to PARPi (Fig. 2a).
Figure 2. Differential sensitivity to PARP inhibition among colon cancer cell lines is not associated with BRCA1 mutation or loss of BRCA1 expression.
(a) Fold change in BARD1β among select colon cancer cell lines were compared using SYBR green real-time PCR and reported as a percentage of the expression level of PARPi sensitive colon cancer cell line, Caco-2. Quantitative PCR was performed at least three times from three different RNA extractions for each cell line. (*p < 0.05 and **p < 0.01) (b) Real-time PCR on polysomal fractions of Caco-2 colon cancer cells showed amplification of various SVs of BARD1. (c) Untreated and 10 μM PARPi treated Caco-2 and SW480 cells for 4 days were stained with both 4′,6-diamidino-2-phenylindole (DAPI) nuclear fluorescent stain and phospho-histone γH2AX DSB marker. Scale bar represents 5 μm. (d) Nuclear phospho-histone γH2AX foci formation in Fig. 3c were quantified (*p < 0.05 and **p < 0.01). (e) Untreated Caco-2 and SW480 cells were stained with DAPI and anti-RAD51 antibody. 10 μM PARPi treated Caco-2 and SW480 cells for 4 days were stained with DAPI and anti-RAD51 antibody. (f) Nuclear RAD51 foci formation in Fig. 2e were quantified (*p < 0.05 and **p < 0.01).
BARD1 SV mRNA is associated with polysomes
To assess potential functions of the BARD1β, we determined whether the SVs mRNAs are translated. Real-time PCR of cDNA with primers targeting the first and last BARD1 exon revealed amplification of multiple SVs in the polysomal fractions, which is congruent with active translation (Fig. 2b).
PARPi causes DSBs in both sensitive Caco-2 and resistant SW480 cells
PARP-1 activity is crucial in the recognition and repair of DNA single-strand breaks. Blocking PARP activity with the PARP-1 inhibitor ABT888 causes single strand breaks to accumulate, leading to formation of double-strand breaks27,16,14. We compared the formation of double-strand breaks in the PARPi-sensitive Caco-2 cells and the PARPi-resistant SW480 and HCT116 cells, using γH2AX foci formation as an indicator (Fig. 2c). Untreated control cells displayed very few or no γH2AX foci formation, consistent with the absence of double-strand breaks, while treatment with the PARPi induced double-strand breaks in both PARPi-sensitive (Caco-2) and PARPi-resistant cells (SW480 and HCT116). PARPi caused significantly more γH2AX staining in the Caco-2 cells (Fig. 2d). This confirms PARPi causes double-strand breaks both in the PARPi-sensitive and PARPi-resistant cell lines further focusing on differences in DNA repair mechanism as the basis for PARPi sensitivity.
PARPi-sensitive colon cancer cells show impaired HR
We hypothesized that BARD1β expression in the PARPi-sensitive colon cancer cells impacts HR proficiency, which can be measured by the formation of RAD51 foci. RAD51 is a critical protein functioning in the HR DNA repair, which localizes to the nucleus to form foci upon DNA damage, and are detected by a florescent antibody and counted using a fluorescent microscope15. We determined the RAD51 foci formation in the PARPi-sensitive and -resistant colon cancer cells following treatment with PARPi. Each cell line was treated with a concentration of PARPi lower than its respective IC50 (10 μM) to avoid lethality. As expected, there were very few or no foci formed in untreated cells (Fig. 2e, left panels; 2f). After treatment with PARPi, PARPi-resistant cells (SW480) (Fig. 2e, right lower panel; 2f) displayed increased RAD51 foci formation compared to PARPi-sensitive colon cancer cells (Caco-2) which displayed a significantly weaker HR DNA repair response characterized by lower RAD51 foci formation (Fig. 2e, right upper panel; 2f). Therefore, impaired HR function may be due to BRCA1/BARD1 deficiency in a BRCA1 wild type setting.
Exogenous expression of SV BARD1β in PARPi-resistant cells results in impaired HR
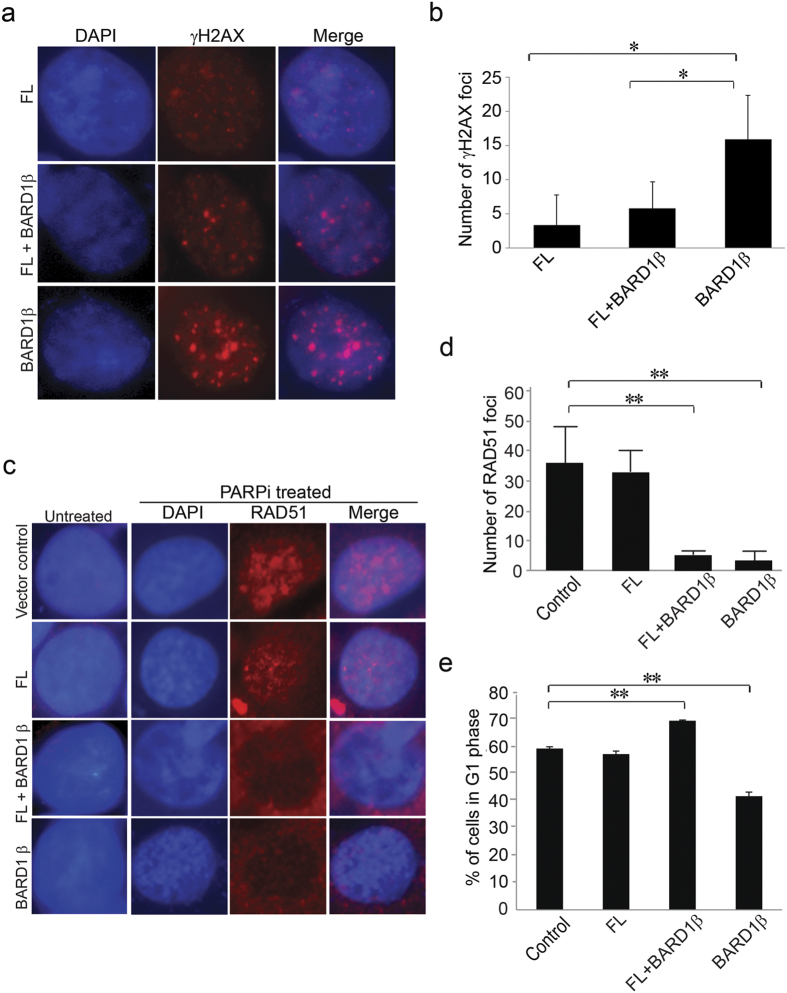
To test the hypothesis that expression of BARD1β leads to impaired HR, we expressed BARD1β in the SW480 PARPi-resistant cells, which have relatively low levels of endogenous BARD1β transcript and have very similar levels of BRCA1 as Caco-2 PARPi-sensitive cells have. Additionally, as reference controls, stable SW480 cells expressing V5 tagged FL BARD1, or both BARD1β and FL BARD1, or an empty vector were prepared (Supplementary Fig. S1). Although HCT116 cells had considerably lower levels of endogenous BARD1β transcript relative to SW480 cells, we decided to prepare stable SW480 cells expressing BARD1β because HCT116 cells have significantly lower levels of endogenous BRCA1 protein, and are deficient in DNA mismatch repair. Exogenous overexpression of BARD1β in SW480 cells caused significantly more γH2AX foci than FL + BARD1β cells in response to PARPi treatment (Fig. 3a,b) indicating increased sensitivity to PARPi.
Figure 3. PARPi-sensitive colon cancer cells show impaired HR.
(a) FL BARD1 expressing SW480 cells, both FL and BARD1β expressing SW480 cells, and BARD1β expressing SW480 cells were exposed to PARPi for 72 hours and stained with DAPI and phospho-histone γH2AX DSB marker. Stably infected SW480 cells were treated with PARPi, and then fixed and stained with DAPI and phospho-histone γH2AX antibody. (b) Nuclear γH2AX foci formation in Fig. 3a were quantified (*p < 0.05 and **p < 0.01). (c) Control SW480 cells, FL BARD1 expressing SW480 cells, both FL and BARD1β expressing SW480 cells, and BARD1β expressing SW480 cells were stained with DAPI and anti-RAD51 antibody. Stably infected SW480 cells were treated with 10 μM PARPi for 4 days, and then fixed and stained with DAPI and anti-RAD51 antibody. (d) Nuclear RAD51 foci formation in Fig. 3c were quantified (*p < 0.05 and **p < 0.01). (e) Control and FL, FL + BARD1β and BARD1β expressing SW480 cells were stained with PI, and monitored by flow cytometry to determine the percentage of G1 phase cells. Experiments were repeated at least three times (*p < 0.05 and **p < 0.01 relative to control).
To evaluate the effect of BARD1β on HR function, RAD51 foci formation was compared among the PARPi-resistant SW480 colon cancer cell lines: SW480 expressing BARD1β, FL BARD1, or both BARD1β and FL BARD1 with and without PARPi treatment (Fig. 3c). In the absence of PARPi, no or only minimal RAD51 foci formation was observed in all cell lines (Fig. 3c, left panels; 3d). However, when the cells were exposed to PARPi, control and FL BARD1 expressing cells displayed significantly higher accumulation of RAD51 (Fig. 3c, left upper three panels; 3d), while both BARD1β expressing and the combined BARD1β and FL BARD1 expressing cells exhibited significantly fewer foci (Fig. 3c, left lower three panels; 3d) consistent with loss of BRCA1/BARD1 function with expression of BARD1β independent of FL BARD1 expression. The reduction of RAD51 foci formation in BARD1β expressing cells does not stem from arrest of cells in G1 phase (Fig. 3e) and (Supplementary Fig. S2). Cell cycle analysis showing no accumulation of cells in G1 phase supports the idea of HR deficiency.
Expression of BARD1β in PARPi-resistant cells increases cytoplasmic localized BRCA1
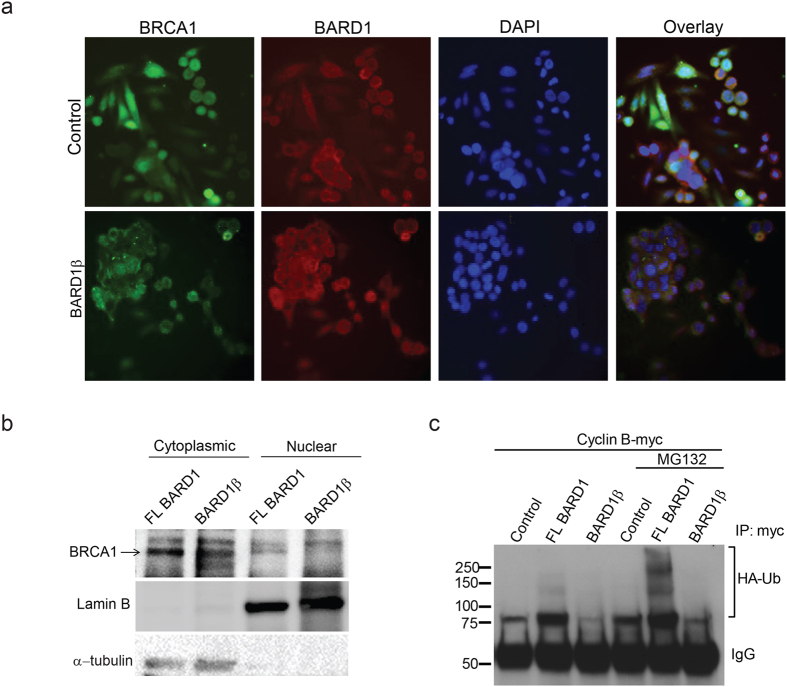
BARD1 plays an important nuclear chaperone role for BRCA128,29,30,31,32,33,34, suggesting that dimerization of BRCA1 with BARD1 is required for nuclear localization of BRCA1. Since BARD1β is missing most of the dimerization region, we examined the subcellular localization of BRCA1 in control and BARD1β stably expressing PARPi-resistant cells using immunofluorescence (Fig. 4a). In control or FL BARD1 overexpressing PARPi-resistant cells, BCRA1 was mainly localized to the nucleus, however, when BARD1β was overexpressed, we observed BRCA1, both, in the cytoplasm and nucleus implying BARD1β is deficient in nuclear chaperone activity (Fig. 4a). Cell fractionation to separate the nuclear and cytoplasmic fractions was performed to confirm differential localization of BRCA1 between FL and BARD1β expressing cells. FL BARD1 expressing cells displayed greater amount of nuclear BRCA1 than BARD1β expressing cells (Fig. 4b).
Figure 4. Expression of BARD1β in PARPi-resistant cells increases cytoplasmic localized BRCA1 and decreased ubiquitin ligase activity of BRCA1.
(a) Control and BARD1β expressing SW480 cells were incubated with anti-BARD1 (red) and anti-BRCA1 (green) for 16 h following fluorescence secondary antibody staining. Nuclei were stained with DAPI. (b) Nuclear and cytoplasmic compartments were isolated by fractionation of stable FL BARD1 and BARD1β expressing SW480 cells. Immunoblotting for BRCA1 is indicated in the top panel, lamin B (control for nuclear fraction) and α-tubulin (control for cytoplasmic fraction) are in the middle and bottom panels respectively. (c) Stable control, full length BARD1 expressing and BARD1β expressing SW480 cells transfected with ubiquitin-FLAG and cyclin B-MYC, and treated with MG132. Detection of ubiquitin-FLAG or cyclin B-MYC was carried by immunoprecipitation with an antibody against MYC followed by immunoblot with an antibody against FLAG-tag.
Impact of BARD1β on ubiquitin ligase activity of BRCA1
Mutations within the RING binding domain leads to impairment of E3 ubiquitin ligase activities of BRCA18,29,30,31. BRCA1 has been reported to ubiquitinate cell cycle proteins including cyclin B; which ultimately contributes to regulation of G2/M cell cycle checkpoint6. We examined the impact of BARD1β expression in PARPi-resistant cells on E3 ubiquitin ligase activity of BRCA1, and ubiquitination of cyclin B. PARPi-resistant cells were co-transfected with His-tagged ubiquitin and MYC tagged cyclin B and treated with MG132, an inhibitor of the proteasome to reduce degradation of ubiquitinated molecules. Consistent with functional BARD1 in control and FL BARD1 overexpressing cells, we detected relatively high ubiquitination activity. In contrast, cells expressing BARD1β displayed little or no ubiquitinated cyclin B following MG132 treatment (Fig. 4c). Taken together, these findings suggest that colon cancer cells which express BARD1β display decreased ubiquitination and are more sensitive to PARPi due to impaired HR.
Expression of BARD1β is associated with a more invasive phenotype
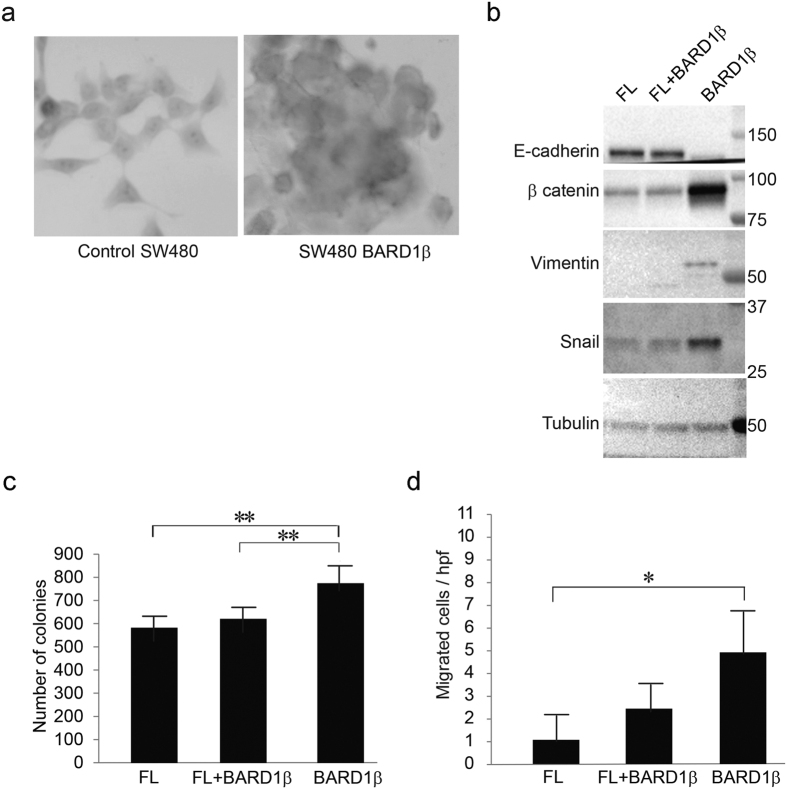
We have previously observed that expression of BARD1β is associated with poor prognosis in colon cancer tumors22. To test if expression of BARD1β leads to a more transformed phenotype, we examined the differences in histology when BARD1β is over-expressed in PARPi-resistant cells (Fig. 5a). Under standard cell culture conditions, control and FL BARD1 expressing PARPi-resistant cells formed single cell monolayers, while BARD1β expressing cells formed clusters indicative of loss of cell-cell adhesion. The transition to metastatic disease includes epithelial to mesenchymal transitions (EMT) and is associated with an increase in cellular ability to form colonies. We examined BARD1β overexpressing PARPi-resistant cells for expression of EMT marker proteins by immunoblotting, and observed a decrease in E-cadherin and increases in β-catenin, Vimentin and Snail, all consistent with induction of EMT (Fig. 5b).
Figure 5. Expression of BARD1β is associated with a more invasive phenotype.
(a) Control (left) and BARD1β expressing (right) SW480 cells were stained with hemotoxylin, and inspected under a light microscope (400x magnification). (b) Stable SW480 cells infected with either FL BARD1, BARD1β or the combination (FL + BARD1β) were harvested, separated, and immunoblotted for EMT proteins: E-cadherin, β-catenin, vimentin, or snail. Tubulin was used as a loading control. (c) Colony formation assay of stable SW480 cells: 100 cells were seeded into six-well plates and incubated for two weeks. Colonies were fixed and stained with crystal violet (*p < 0.05 and **p < 0.01). (d) The relative number of migrated stable SW480 cells per high power field (hpf) (*p < 0.05 and **p < 0.01).
Next, we compared the clonogenic capacity of PARPi-resistant cells expressing either FL BARD1 or BARD1β, or both using the colony formation assay as previously described32. BARD1β expressing cells produced significantly higher numbers of colonies relative to cells expressing either FL or both FL BARD1 and BARD1β, indicating that BARD1β expressing cells retain a higher capacity to produce colonies (Fig. 5c). Increase in migration of BARD1β expressing cells further supports the notion of enhanced tumorigenesis (Fig. 5d).
Exogenous expression of SV BARD1β in PARPi-resistant colon cancer cells imparts sensitivity to PARP inhibition
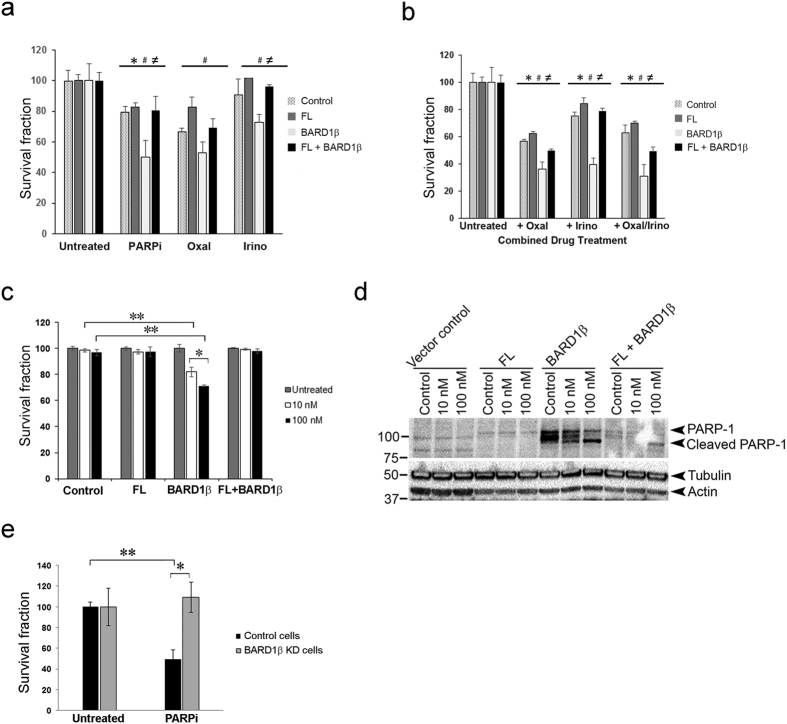
To evaluate the effect of BARD1β expression on PARPi-mediated cell death, we exposed stable transfectants of PARPi-resistant cells, characterized above, to low dose PARPi. Compared to control and FL BARD1 expressing cells, expression of BARD1β sensitized cells to low concentrations of PARPi (Fig. 6a). When FL BARD1 and BARD1β are co-expressed, we observed an intermediate phenotype with respect to PARPi sensitization (Fig. 6a).
Figure 6. Exogenous expression of SV BARD1β in resistant SW480 cells imparts sensitivity to PARPi.
(a) SW480 cells stably infected with either FL BARD1, BARD1β, or the combination FL + BARD1β, were exposed to either PARPi (20 μM) for 96 hours, or only Oxaliplatin (50 μM) for 36 hours, or only Irinotecan (5 μM) for 36 hours. Survival fraction was determined using cell viability assays (*control vs. BARD1β; #FL vs. BARD1β; ≠BARD1β vs. FL + BARD1β; *,#,≠p < 0.01). (b) Stable SW480 cells which were prepared as explained above were exposed to PARPi (20 μM) for 84 h followed by either Oxaliplatin (50 μM) or Irinotecan (5 μM) or both supplementation for 36 h. Survival fraction was determined using cell viability assays (*control vs. BARD1β; #FL vs. BARD1β; ≠BARD1β vs. FL + BARD1β; *,#,≠p < 0.01). (c) SW480 cells stably infected with either FL BARD1, BARD1β, or the combination FL + BARD1β, were exposed to Olaparib PARPi (10 nM or 100 nM) for 72 hours. Survival fraction was determined using cell viability assays (*p < 0.05 and **p < 0.01). (d) Protein lysates (40 μg) from each sub-line of SW480 cells were separated and immunoblotted with anti-PARP1 antibody as described in Methods. For normalization, blots were probed with α-tubulin and β-actin. This blot is representative of three independent experiments. (e) shRNAs targeting BARD1β were prepared, and BARD1β was stably knocked down in Caco-2 cells as described in Methods. Cells were exposed to 10 μM ABT-888 for 72 hours. Survival fraction was determined using cell viability assays (*p < 0.05 and **p < 0.01).
DNA damaging agents including Irinotecan and Oxaliplatin are commonly used in the treatment of metastatic colorectal cancers33. Irinotecan is a topoisomerase 1 poison leading to the formation of a potentially lethal DNA double-stranded break. Oxaliplatin, in contrast, is a platinum analogue, which causes the formation of inter- and intra-strand crosslinks in DNA that prevent transcription and replication. Ultimately this results in DNA damage and cell death. We investigated if PARPi can sensitize colon cancer cells expressing BARD1β in vitro to either Oxaliplatin and/or Irinotecan treatment. We established the optimal time of drug exposure and observed that Irinotecan and Oxaliplatin work more rapidly and achieve optimal action at 36 hours while PARPi requires 96 hours of incubation (Supplementary Fig. S3). Similarly we established concentrations for each drug that are sub-lethal to allow for optimal action when used in combination (Supplementary Fig. S3). Expression of BARD1β in PARPi-resistant cells did not selectively sensitize the cells to either Oxaliplatin or Irinotecan alone relative to control cells (Fig. 6a). Due to the differential time course of drug response, we chose to treat cells sequentially, first with PARPi followed by Oxaliplatin or Irinotecan. Addition of Oxaliplatin or Irinotecan decreased the fraction of viable cells in all stable PARPi-resistant cells (Fig. 6b); however, addition of Irinotecan to PARPi pre-treated cells created higher specificity with respect to killing of BARD1β expressing PARPi-resistant cells. These data indicate that colon cancer patients with high expression of BARD1β may receive therapeutic benefit from a serial combination of PARP inhibition and Irinotecan treatment.
To test whether BARD1β expressing SW480 cells was exclusively sensitive to ABT-888 or other PARP inhibitors display a similar feature, we used a potent PARP inhibitor, Olaparib (AZD2281), which has the additional cytotoxic function due to its higher ability of PARP-trapping on chromatin. Consistent to our findings with ABT-888, BARD1β expression in SW480 cells increased sensitivity to Olaparib (Fig. 6c) as well.
In addition to these survival assays, we evaluated whether exogenous expression of BARD1β in SW480 cells increased the overall apoptotic index. Expression of BARD1β increased PARP1 protein expression. Moreover, cleavage of PARP1 was further stimulated after 10 nM and 100 nM treatments of Olaparib which was an indication of increased apoptosis (Fig. 6d).
To further confirm the expression of BARD1β is associated with PARPi sensitivity, we knocked down the expression of BARD1β in PARPi sensitive Caco-2 cells using shRNA-mediated knockdown. Knockdown of BARD1β in Caco-2 cells rendered these cells less sensitive to PARPi relative to control Caco-2 cells (Fig. 6e).
In summary, we show that colon cancer cells which express BARD1β SV display impaired HR DNA repair and are more sensitive to PARP-1 inhibition. Overexpression of BARD1β in PARPi-resistant colon cancer cells rendered these cells sensitive to PARPi, and decreased E3 ligase activity as well as nuclear localization of BRCA1. This was associated with an increase in EMT, as well as colony formation and enhanced cell migration, supporting that oncogenic properties of BARD1β in colon cancer are at least in part, via targeting BRCA1 function.
Discussion
The tumor suppressor functions of BRCA1 are largely attributed to stabilization of BRCA1 by heterodimer formation with binding partners such as BARD1 within the BRCA1-associated RING domain protein. About twenty percent of clinically relevant BRCA1 mutations occur within the binding site of BRCA1 and BARD1, indicating that the binding of BARD1 is crucial for HR DNA repair function of BRCA15,34. BRCA1 is typically wild type in colon cancer, but the presence of various BARD1 SVs of mostly unclear function has been reported7,22,23.
The loss of BARD1 in mice leads to embryonic lethal phenotype and chromosomal instability9,35. BARD1 spans 11 exons and we previously observed 19 distinct BARD1 SVs in colon cancer patients that account for over 40 percent of the total BARD1 transcript22. The individual functions of these SVs have not been assessed. However, BARD1β may have oncogenic activities. In cervical, breast, ovarian and endometrial cancer cell lines, BARD1 SVs missing the RING domain have been reported to be more abundant than the full length protein, and the knockdown of both FL and SVs decreased the rate of proliferation and led to growth arrest25.
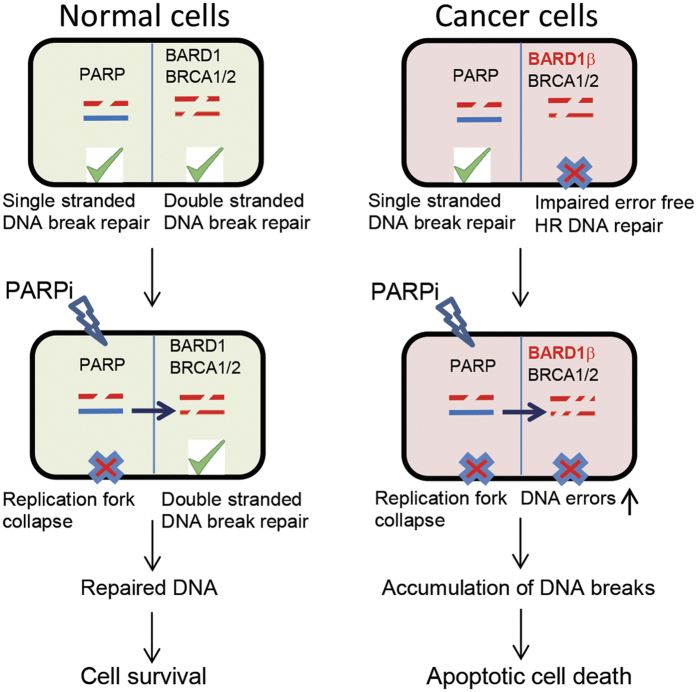
Deficiencies in HR DNA repair sensitize cancer cells to PARPi36. PARPi leads to the accumulation of single-strand DNA breaks which are converted to double-strand DNA breaks. In normal cells, PARPi alone is not sufficient to induce apoptosis because the HR DNA repair mechanism repairs double-strand DNA breaks (Fig. 7) leading to cell survival. However, in cancer cells with HR deficiency, other error prone DNA repair mechanisms dominate which increases DNA errors and genomic instability, and ultimately induces apoptosis14. We found that BRCA1 protein expression in SW620, Caco-2, and SW480 cells was similar; however, Caco-2 cells had significantly higher levels of BARD1β transcript. This observation supports the idea of BARD1β expression level is a marker for PARPi sensitivity. Overexpression of BARD1β in colon cancer cell lines, such as in Caco-2 cells, impairs error free HR DNA repair by negatively influencing functions of BRCA1 thus sensitizing cells to PARPi. The high numbers of DNA breaks caused by HR DNA repair deficiency coupled with PARP inhibition leads to massive genomic instability resulting in apoptotic cancer cell death (Fig. 7). This increased sensitivity to the PARPi, suggests therapeutic applications for cancer patients expressing this BARD1 isoform. An effective ex vivo assay to determine the proficiency of HR in colon cancer patients will be useful for predicting which patient might obtain the most benefit from the therapeutic use of PARPi. Response to PARPi therapy could be predicted using the ex vivo RAD51 foci formation assay37. This would ultimately provide a biologic basis for therapeutic strategies such as PARPi alone or combination of PARPi with DNA damaging agents in the treatment of colon cancer patients.
Figure 7. Schematic of sensitization of colon cancer cells to PARPi via BARD1β expression.
In normal cells, single strand DNA breaks are repaired via PARP-1 while double strand DNA breaks are repaired via BRCA1/BARD1 dependent HR repair. PARPi treatment of normal cells leads to replication fork collapse and persistence of single strand DNA breaks. However, single strand breaks are converted to double strand DNA breaks by replication and then repaired by HR resulting in cell survival. Therefore, HR DNA repair proficient normal cells are resistant to PARP-1 inhibition. In contrast, overexpression of BARD1β decreases the rate of error-free HR of double strand DNA breaks. HR DNA repair deficiency renders these tumor cells hypersensitive to PARPi leading to apoptosis.
In this study, we demonstrate that overexpression of BARD1β stimulated a decrease in BRCA1 functional capabilities with decreased ubiquitination, decreased nuclear translocation, and decreased HR repair. These results indicate that expression of BARD1 SVs lacking the BRCA1 binding domain could predispose patients to colon cancer similar to the predisposition for breast cancer by BRCA1 mutations. We have previously reported that BARD1β is overexpressed in a subset of colorectal cancer patients. Due to the lack of the ring binding domain, BARD1β does not contain the binding site necessary for interaction with BRCA1 and may display oncogenic effect via affecting BARD1/BRCA1 tumor suppressor function either independent or dependent on FL BARD1 protein. For example, Ryser et al. report that FL BARD1 binds to BRCA1 leading to ubiquitination and degradation of Aurora B during mitosis. In contrast BARD1 SV can bind the Aurora B/BRCA2 complex stabilizing Aurora B leading to enhanced cell proliferation38. Additional binding partners, such as FL BARD1 and/or PARP, may exist for this BARD1 isoform which contribute to its oncogenic effect. We found that BARD1β expressing SW480 cells had higher levels of PARP1 protein expression relative to FL or both FL + BARD1β expressing SW480 cells. It has been previously reported that PARP1 also has a role in repair of double strand breaks, such as non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ), which are error prone DNA repair pathways39,40, therefore overexpression of PARP1 in BARD1β expressing cells might also increase genomic stability and cancer.
It has been previously reported that while overexpression of BARD1 SVs results in cell transformation, its repression leads to growth arrest of cancer cells25. The exogenous expression of BARD1β isoform in PARPi-resistant SW480 colon cells leads to a more transformed phenotype with enhanced EMT. E-cadherin, a mediator of cell-cell adhesion41, is reduced by BARD1β overexpression which can support invasion and growth of tumor cells. In contrast, β-catenin expression was markedly higher in BARD1β expressing cells. β-catenin overexpression has been linked to many epithelial cancers, including colorectal cancer42. BARD1β expressing cells exhibited increased levels of the EMT regulator, SNAIL, and mesenchymal marker, vimentin, which could contribute to the observed histological difference between control and BARD1β expressing cells.
In conclusion, BARD1β displays oncogenic activities through negatively affecting the tumor suppressor functions of BRCA1. Our findings indicate therapeutic targeting of deficient HR with PARPi therapy may be beneficial to the colon cancer patients who express this SV. Future development of fast, novel detection methods to identify colon cancer patients with HR deficiency is needed and detection of BARD1β may provide direction.
Methods
Chemicals
Aqueous solutions were prepared of ABT888 (sc-202901, Santa Cruz Biotechnology, Dallas, TX), stock concentration of 2 mM; Oxaliplatin (O9512, Sigma, St. Louis, MO), stock concentration of 10 mM; Irinotecan (I1406, Sigma), stock concentration of 10 mM; and Olaparib (10621, Cayman, Ann Arbor, MI), stock concentration of 10 mM.
Cell culture and generation of stable cell lines
SW480 and Caco-2 cells (ATCC, Manassas, VA, USA) were maintained in 10% Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic agent in a 37 °C incubator with 5% CO2. Cells were validated by 9 STR (short tandem repeat) profiling, using CellCheck 9 Plus, tested for mycoplasma, and found to be mycoplasma free (both assays performed by IDEXX, Columbia, MO) in February 2015. BARD1β and FL BARD1-expressing lentivirus were constructed in pCDH-CMV-MCS-EF1-Puro cDNA cloning and expression vector (CD510B-1, System Biosciences, San Francisco, CA, USA). SW480 cells stably overexpressing BARD1β, FL BARD1, or both BARD1β and FL BARD1 were generated using lentivirus infection.
Immunoblotting
Immunoblotting was performed using the Trans-blot Turbo transfer system (Bio-Rad, Hercules, CA) using PVDF membranes. Membranes were then incubated with anti-BRCA1 (Abcam, Cambidge, MA), Flag antibody (Sigma-Aldrich, St. Louis, MO), V5 tag, E-cadherin, snail, vimentin, or β-catenin (Cell Signaling, Danvers, MA), cleaved PARP1 (Santa Cruz, Dallas, TX) primary antibodies for 16 hr at 4 oC.
Immunofluorescence
Cells seeded on glass coverslips were fixed in 4% paraformaldehyde, followed by blocking with 3% BSA in PBS. Then, cells were incubated with anti-RAD51 (Santa Cruz), anti-P-histone H2AX (Cell Signaling), anti-BRCA1 (Abcam), or anti-BARD1 (Santa Cruz) antibody in PBS, followed by incubation with goat anti-rabbit and anti-mouse IgG conjugated with Alexa Fluor 488 and 594 (Invitrogen) in PBS. Cells were washed in PBS, mounted, and then imaged with a fluorescence microscope (Nikon Eclipse E600 microscope, SPOT Imaging Software, Diagnostic Instruments, MI). γH2AX and RAD51 immunofluorescence assays were performed as previously described36,43,44. An average of 100 nuclei were counted for γH2AX and RAD51 immunofluorescence assays, and experiments performed in triplicates.
Measurement of proliferation rates and cell survival
FET, HCT116 p21−/−, HCT116, SW480, SW620 and Caco-2 cells were seeded into 96-well plate at a concentration of 20,000 cells with 100 μL/well DMEM supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic agent in a 37 °C incubator with 5% CO2 and incubated for 24 hr. A range of concentrations (0, 1, 5, 10, 20, 40 μM) of PARPi were added to the seeded cells and incubated for 72 or 96 hr. Cell proliferation and cell survival were determined according to manufacturer’s instructions using either Cell Counting Kit-8 from Dojindo Laboratories (Rockville, MD) and/or the BioRad automated cell counter (Bio-Rad). The half maximal inhibitory concentrations (IC50) of PARPi were determined by constructing a dose-response curve for each cell line. The experiment was performed at least three independent trials with at least three technical replicates.
Migration assay
Migration assays were performed as previously described45 using transwell 12 well plates with 8 μm pores (Corning, Tewksbury, MA) coated with fibronectin (Sigma). Data are presented as the number of migrated cells per high power field (hpf) as the mean +/− s.d. of triplicate assays.
Ubiquitination assay
E3 ubiquitination ligase activity of BARD1 proteins were compared in control, FL BARD1, and BARD1β expressing SW480 cells as previously described46. These cells were co-transfected with HA-tagged ubiquitin and MYC-tagged cyclin B using Fugene HD in the presence and absence of 10 μM proteasome inhibitor MG132 for 3 hr. After 48 hr, Myc-tagged cyclin B proteins were immunoprecitated with anti-Myc antibody, and resolved in 10% SDS PAGE. PVDF membrane was immunoblotted with HA antibody.
Polysomal fraction assay
To confirm translation of BARD1 SVs, we isolated polysomal fractions from Caco-2 cells. Cells were treated with 0.1 mg/ml cycloheximide (CHX) for 3 min at 37 °C, and harvested into hypotonic lysis buffer [0.5% Triton X-100, 0.5% sodium deoxycholate, 2.5 mM MgCl2, 5 mM Tris (pH 7.5), 1.5mM KCl, 0.1 mg/ml CHX, 100 units RNAse-Inhibitor (Ambion-Life Technologies, Grand Island, NY)] at 4 °C. Nuclei were removed, 200 μg/ml heparin was added and residual debris was removed by centrifugation. The lysate was layered on a 10 ml continuous sucrose gradient (10–50% sucrose in 5 mM MgCl2, 20 mM HEPES (pH 7.6), 100 mM KCl). After 110 min of centrifugation at 35.000 rpm at 4 °C, the absorbance at 254 nm was measured continuously as a function of gradient depth using the Teledyne Isco UA-6 Absorbance Detector. Prepared polysomal fractions were pooled, and total RNA from polysomal fractions was isolated with the Allprep RNA/Protein Kit from Qiagen (Hilden, Germany). Reverse transcription-PCR of isolated RNA was performed with the primer pair targeting the first and last exon of the BARD1 transcript, and the PCR products were visualized on a 1% w/v agarose gel stained with ethidium bromide as previously described22.
Quantitative real-time PCR
To quantify the expression of BARD1β, SYBR green quantitative real-time PCR reaction was performed as previously described22 with the following modifications. Superscript™ III First-Strand Synthesis SuperMix and Oligo(dT)20 primers by Invitrogen (Carlsbad, CA) was used for cDNA synthesis. The expression of BARD1β was normalized to GAPDH and the results were analyzed by the comparative Ct method. Quantitative PCR was performed at least three times from three different RNA extractions for each cell line. Statistical comparisons were performed on mean expression of the independent samples and fold differences among colon cancer cell lines were calculated.
Cell cycle analysis
The cell cycle distribution of stable SW480 cells was analyzed by using propidium iodide (PI) staining. Briefly, 106 cells were trypsinized and washed with PBS, centrifuged and fixed in ice-cold 70% ethanol and washed with cold PBS, and supernatant was removed. For measurement of DNA content, cells were washed with PBS and then stained with 100 μg/mL RNase A in PBS (Sigma), 50 μg/mL PI (Sigma) and 0.1% (v/v) Triton X-100 for 30 min at room temperature in dark, and analyzed by flow cytometry (Becton-Dickinson, Franklin Lakes, NJ, USA) with Cell Quest software. Three independent experiments were performed for each cell line.
Gene knockdown
We conducted shRNA-mediated knockdown of BARD1β gene in Caco-2 cells and studied the effects on cell viability. The BARD1b target sequence was: (5′-GCTAGCCACTGCTCAGTAATG). We constructed short hairpin RNA (shRNA) oligos against BARD1β and cloned into pLKO.1 puro lentiviral backbone according to Addgene’s pLKO.1 protocol. Lentiviral particles were generated in 293T cells and transduced into Caco-2cells.
Statistical analysis
All data presented as mean +/− SD. One-way ANOVA and Tukey’s multiple comparison test were used as appropriate comparison among groups. The criterion for statistical significance was set at *p < 0.05 or **p < 0.01.
Additional Information
How to cite this article: Ozden, O. et al. Expression of an Oncogenic BARD1 Splice Variant Impairs Homologous Recombination and Predicts Response to PARP-1 Inhibitor Therapy in Colon Cancer. Sci. Rep. 6, 26273; doi: 10.1038/srep26273 (2016).
Supplementary Material
Acknowledgments
Support for this research was provided by a National Institutes of Health grant R01CA141057 to B.J., and Martha Hofmann Levin and Anna C. Hofmann Scholar award, Institutional NRSA, clinical oncology research training program, No. T32 CA079447 to F.B. We thank Northwestern SDRC, DNA/RNA Delivery core facility for the generation of stable cells.
Footnotes
Author Contributions O.O. and F.B. designed the study, performed majority of the experiments, performed analysis, interpreted data. O.O., N.K. and B.J. wrote the manuscript. O.O. and J.B. made figures for the manuscript. S-H.P., A.J. and J.C.S. performed part of the experiments. S.H.B., J.J.S., J.B. and C.Y. assisted part of the experiments. All authors reviewed the manuscript. B.J. is the guarantor and gave the final decision to submit this manuscript for publication.
References
- Situ B. C. I. Cancer Facts & Figures 2015. American Cancer Society. Available at: http://www.cancer.org/acs/groups/content/@editorial/documents/document/acspc-044552.pdf (Accessed: 24th March 2016).
- Weitz J. et al. Colorectal cancer. Lancet 365, 153–165 (2005). [DOI] [PubMed] [Google Scholar]
- SEER Cancer Statistics Factsheets: Colon and Rectum Cancer. National Cancer Institute. Bethesda, MD. Available at: http://seer.cancer.gov/statfacts/html/colorect.html (Accessed: 24th March 2016).
- Tennstedt P. et al. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int J Cancer 132, 2118–2126 (2013). [DOI] [PubMed] [Google Scholar]
- Ohta T., Sato K. & Wu W. W. The BRCA1 ubiquitin ligase and homologous recombination repair. Febs Letters 585, 2836–2844 (2011). [DOI] [PubMed] [Google Scholar]
- Shabbeer S. et al. BRCA1 targets G2/M cell cycle proteins for ubiquitination and proteasomal degradation. Oncogene 32, 5005–5016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. J. C. et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genetics 14, 430–440 (1996). [DOI] [PubMed] [Google Scholar]
- Hashizume R. et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. Journal of Biological Chemistry 276, 14537–14540 (2001). [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I. & Jefford C. E. Is there more to BARD1 than BRCA1? Nature Reviews Cancer 6, 382–391 (2006). [DOI] [PubMed] [Google Scholar]
- Miki Y. et al. A Strong Candidate for the Breast and Ovarian-Cancer Susceptibility Gene Brca1. Science 266, 66–71 (1994). [DOI] [PubMed] [Google Scholar]
- Savage K. I. & Harkin D. P. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. Febs J 282, 630–646 (2014). [DOI] [PubMed] [Google Scholar]
- Ghimenti C. et al. Germline mutations of the BRICA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes & Cancer 33, 235–242 (2002). [DOI] [PubMed] [Google Scholar]
- Phelan C. M. et al. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: results from a follow-up study. Br J Cancer 110, 530–534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy. Nucleic Acids Res 42, 6106–6127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- Farmer H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- Fong P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- Arnaudeau C., Lundin C. & Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. Journal of Molecular Biology 307, 1235–1245 (2001). [DOI] [PubMed] [Google Scholar]
- Li M. & Yu X. C. Function of BRCA1 in the DNA Damage Response Is Mediated by ADP-Ribosylation. Cancer Cell 23, 693–704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. D. et al. PARP1-Driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination-Mediated DNA Repair. Cancer Discovery 4, 1430–1447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman F. E. et al. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev 19, 1227–1237 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn J. C., Hothorn T. & Jung B. BARD1 Expression Predicts Outcome in Colon Cancer. Clinical Cancer Research 17, 5451–5462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Q. et al. Expression of oncogenic BARD1 isoforms affects colon cancer progression and correlates with clinical outcome. British Journal of Cancer 107, 675–683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse K. R. et al. Common Variation at BARD1 Results in the Expression of an Oncogenic Isoform That Influences Neuroblastoma Susceptibility and Oncogenicity. Cancer Research 72, 2068–2078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Research 67, 11876–11885 (2007). [DOI] [PubMed] [Google Scholar]
- Lord C. J. & Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med 19, 1381–1388 (2013). [DOI] [PubMed] [Google Scholar]
- McGlynn P. & Lloyd R. G. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol 3, 859–870 (2002). [DOI] [PubMed] [Google Scholar]
- Fabbro M., Rodriguez J. A., Baer R. & Henderson B. R. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem 277, 21315–21324 (2002). [DOI] [PubMed] [Google Scholar]
- Chen A., Kleiman F. E., Manley J. L., Ouchi T. & Pan Z. Q. Autoubiquitination of the BRCA1-BARD1 RING ubiquitin ligase. Journal of Biological Chemistry 277, 22085–22092 (2002). [DOI] [PubMed] [Google Scholar]
- Jefford C., Feki A., Harb J., Irminger-Finger E. T. & Krause K. Characterization of BARD1 protein regions correlated with sub-cellular localization and required for apoptotic activity. Mol Biol Cell 13, 306a–306a (2002). [Google Scholar]
- Jefford C. E., Feki A., Harb J., Krause K. H. & Irminger-Finger I. Nuclear-cytoplasmic translocation of BARD1 is linked to its apoptotic activity. Oncogene 23, 3509–3520 (2004). [DOI] [PubMed] [Google Scholar]
- Franken N. A. P., Rodermond H. M., Stap J., Haveman J. & van Bree C. Clonogenic assay of cells in vitro. Nature Protocols 1, 2315–2319 (2006). [DOI] [PubMed] [Google Scholar]
- Rao G. H. et al. Establishment of a human colorectal cancer cell line P6C with stem cell properties and resistance to chemotherapeutic drugs. Acta Pharmacol Sin 34, 793–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. & Weiss W. A. Alternative splicing in cancer: implications for biology and therapy. Oncogene 34, 1–14 (2015). [DOI] [PubMed] [Google Scholar]
- McCarthy E. E., Celebi J. T., Baer R. & Ludwig T. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Molecular and Cellular Biology 23, 5056–5063 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A. et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res 16, 2344–2351 (2010). [DOI] [PubMed] [Google Scholar]
- Shah M. M. et al. An ex vivo assay of XRT-induced Rad51 foci formation predicts response to PARP-inhibition in ovarian cancer. Gynecol Oncol 134, 331–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser S. et al. Distinct Roles of BARD1 Isoforms in Mitosis: Full-Length BARD1 Mediates Aurora B Degradation, Cancer-Associated BARD1 beta Scaffolds Aurora B and BRCA2. Cancer Research 69, 1125–1134 (2009). [DOI] [PubMed] [Google Scholar]
- Wang M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34, 6170–6182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. et al. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis 6, e1697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J. & Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Bio 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. J. et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275, 1787–1790 (1997). [DOI] [PubMed] [Google Scholar]
- Lee S. A., Roques C., Magwood A. C., Masson J. Y. & Baker M. D. Recovery of deficient homologous recombination in Brca2-depleted mouse cells by wild-type Rad51 expression. DNA Repair (Amst) 8, 170–181 (2009). [DOI] [PubMed] [Google Scholar]
- Paull T. T. et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10, 886–895 (2000). [DOI] [PubMed] [Google Scholar]
- Bauer J., Sporn J. C., Cabral J., Gomez J. & Jung B. Effects of activin and TGFbeta on p21 in colon cancer. Plos One 7, e39381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizin E. & Irminger-Finger I. Negative feedback loop of BRCA1-BARD1 ubiquitin ligase on estrogen receptor alpha stability and activity antagonized by cancer-associated isoform of BARD1. Int J Biochem Cell Biol 42, 693–700 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.