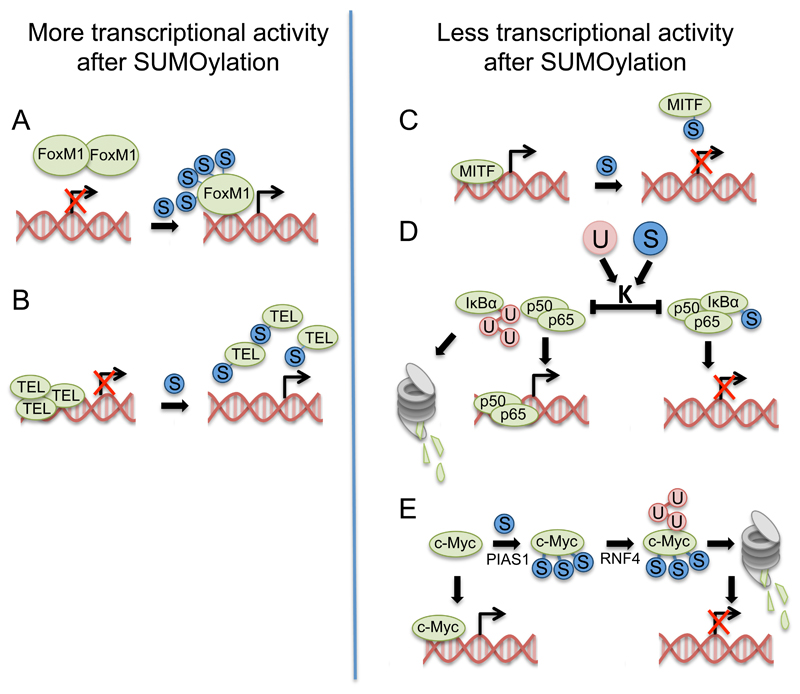
Figure 4. SUMOylation modulates the activity of transcription factors.
The majority of SUMO targets identified so far are involved in transcriptional regulation and chromatin remodelling. The effect of SUMOylation on the target protein can differ greatly and therefore has to be examined for each SUMO target and each SUMO site individually. This figure shows some of the mechanisms identified for transcription factors involved in cancer development. A) SUMO (S in blue circles) is known to affect protein-protein interactions and can therefore either obstruct binding to transcriptional inhibitors or promote the formation of inhibitory protein complexes. SUMOylation of FoxM1, for example, has been described to inhibit the formation of protein dimers and thereby induce transcriptional activity [23]. B) Similar to FoxM1, SUMOylation of the transcriptional repressor TEL/ETV6 blocks the formation of the multimer, which is needed to repress transcriptional activity at promoter regions. Therefore SUMOylation of TEL also promotes gene expression [85]. C) A SUMO-deficient mutant of MITF shows enhanced binding to DNA and increased expression of the target gene HIF1α. Therefore it has been suggested that SUMOylation of MITF blocks DNA binding and leads to decreased transcriptional activity [79] D) SUMOylation can also influence the stability of a target protein. SUMO and ubiquitin (U in red circles) can compete for the same lysine (K) residues as described for IκBα, a repressor of the multimeric transcription factor NFκB. While ubiquitylation of IκBα leads to proteasomal degradation and the release of the two subunits p65 and p50 and therefore enhances transcriptional activity, SUMOylation of IκBα stabilizes the protein and blocks gene expression [105]. E) However, the presence of multiple SUMO moieties can promote ubiquitylation via SUMO targeted ubiquitin ligases (Stubl) as described as a potential mechanism for c-Myc. SUMOylation via PIAS1 promotes ubiquitylation via the Stubl RNF4 and subsequent proteasomal degradation, thereby reducing transcriptional activity [73].