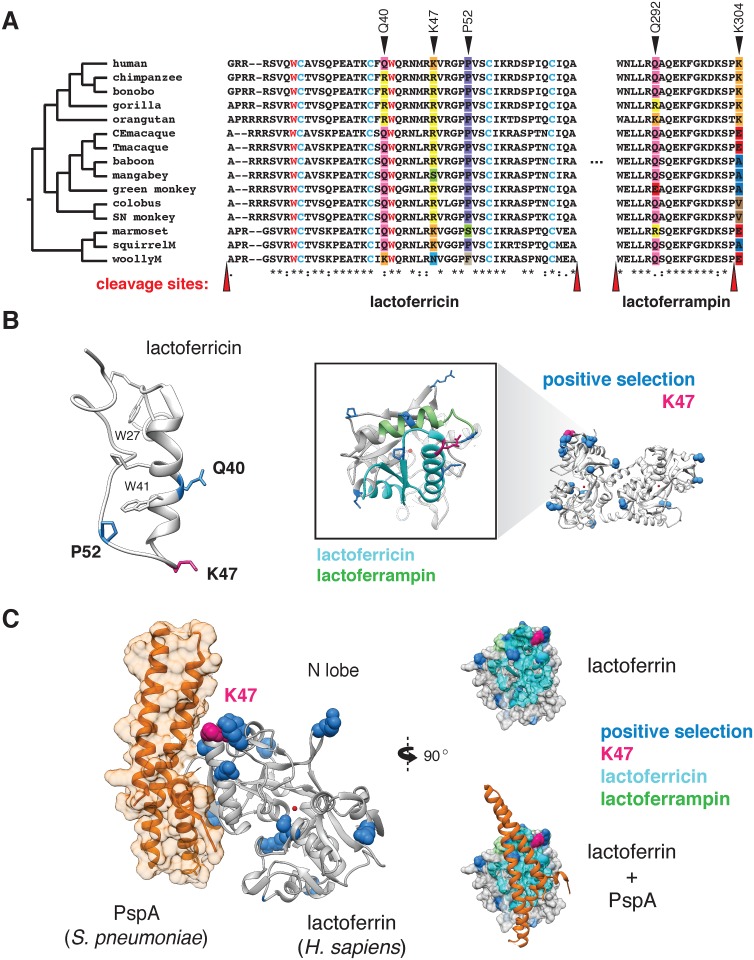
Fig 3. Rapid evolution of lactoferrin-derived antimicrobial peptides and pathogen binding interfaces.
A. Amino acid alignment of the lactoferricin and lactoferrampin peptide sequences across primates. Sites under positive selection are denoted with black arrows, with amino acids at these positions color-coded. Conserved tryptophan (red) and cysteine (blue) residues are highlighted, which contribute to target membrane interactions and disulfide bond formation respectively. The reported cleavage sites of the two peptides are denoted with red arrows. B. Left: solution structure of the free human lactoferricin peptide (PDB: 1Z6V), with sites under positive selection (blue), including position 47 (magenta) indicated. Conserved tryptophan and cysteine residues highlighted in A are also shown. Right: enlarged view of the human lactoferrin N lobe highlighting sequences corresponding to lactoferricin (cyan) and lactoferrampin (green) antimicrobial peptides. Sites previously identified under positive selection in primates are shown in blue, with the position 47 variant shown in magenta. C. Crystal structure (PDB: 2PMS) of human lactoferrin N lobe (gray) bound to PspA from Streptococcus pneumoniae (orange). Side chains of sites under positive selection (blue), including position 47 (magenta) are shown.