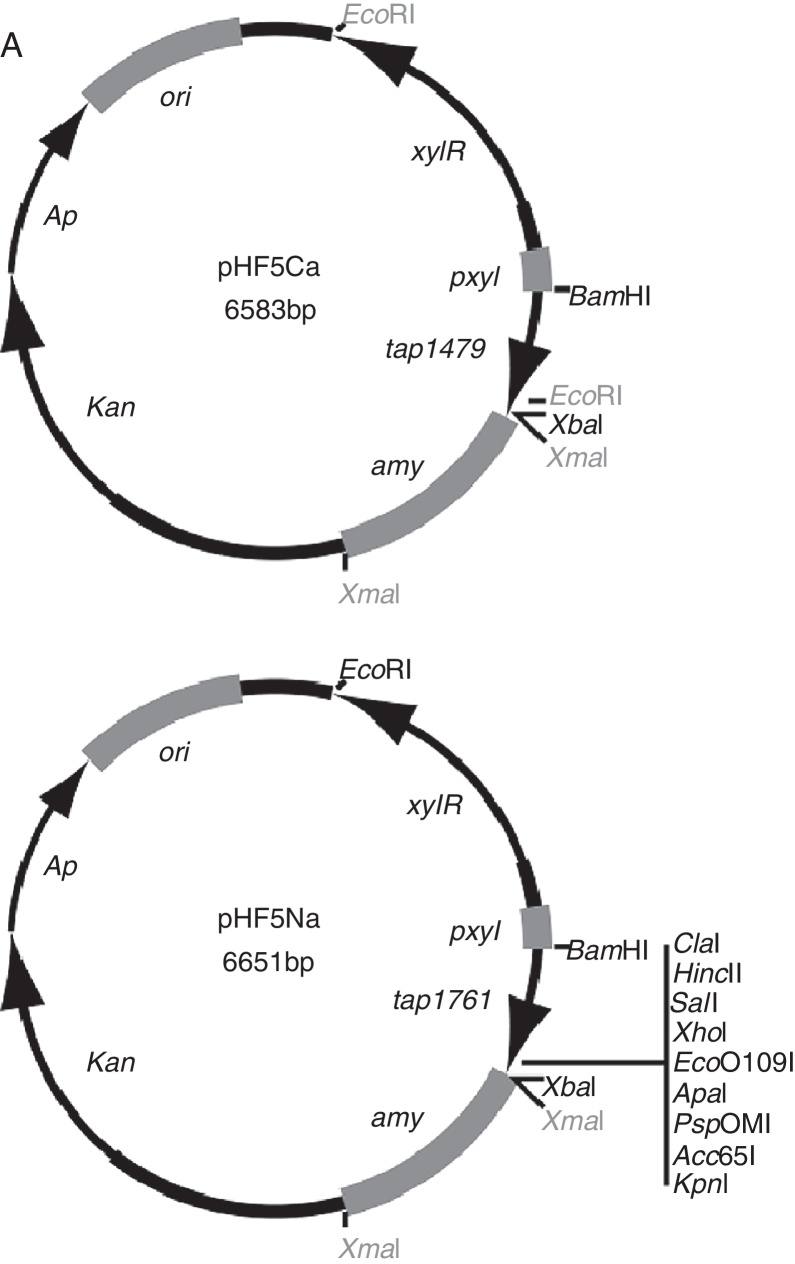
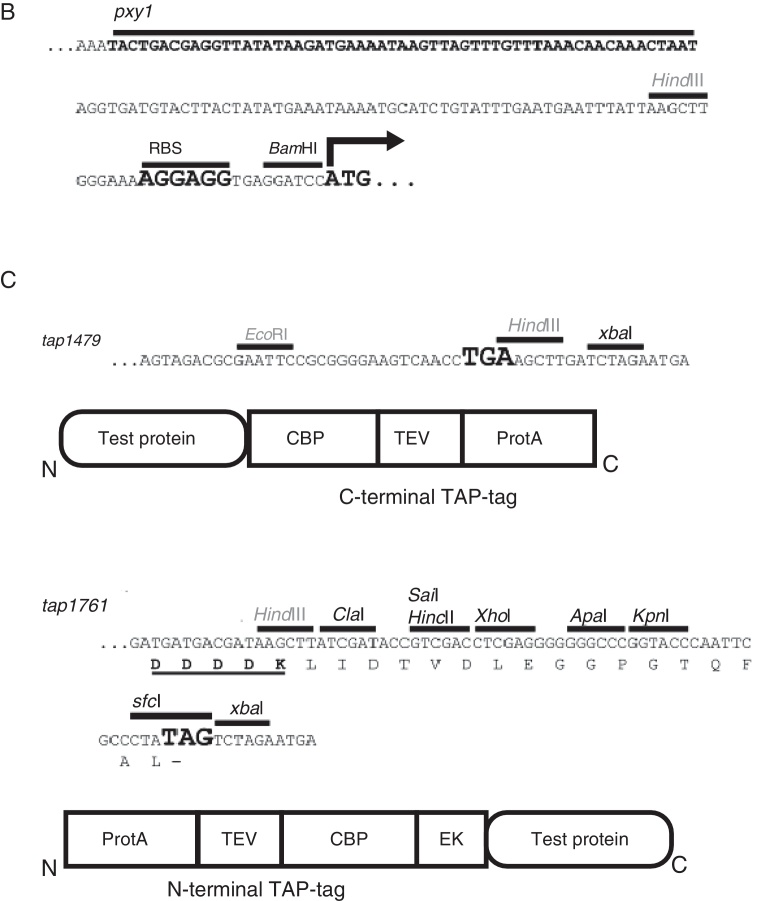
Fig. 1.
Expression vectors for Tandem Affinity purification in Xac. (A) Schematic representation of pHF5Ca and pHF5Na: the protein expression vectors for TAP-tagging in Xac. Each vector carries a different TAP-coding DNA, tap1479 and tap1761 respectively, which produce TAP fusions to either the C- or N-terminal ends of proteins under study. Both vectors are based on the pCR2.1-TOPO backbone which carries a pUC replication origin and DNA cassettes for ampicillin (Ap) and kanamycin (kan) resistance. (B) DNA sequence of the xylose promoter region common to pHF5Ca and pHF5Na showing the location of the ribosome binding-site (RBS) and the start codon (ATG). (C) The DNA sequences of the 3′-ends of tap1479 and tap1761. Underneath each sequence is the schematic view of the protein fusion that can be produced using the tag. Note that the organization of the modules that compose the TAP-tags (CBP, TEV, and ProtA) differ with respect to TAP1479 and TAP1761 (see text); for the C-terminal fusions, the TAP1479 has ProtA located at the extreme C-terminus of the polypeptide, while ProtA is at the beginning of the protein in the N-terminal fusions. Unique restriction sites for DNA cloning are shown in black; non-unique sites in gray. CBP, calmodulin binding-protein; TEV, Tobacco Etch Virus Protease recognition sequence; ProtA, Protein A from Staphylococcus aureus; and EK, enterokinase recognitions sequence (DDDDK, present only in TAP1761); pxyl, xylose promoter; xylR, the xylose repressor gene; amy, an 800 bp fragment of the alpha-amylase gene from Xac.