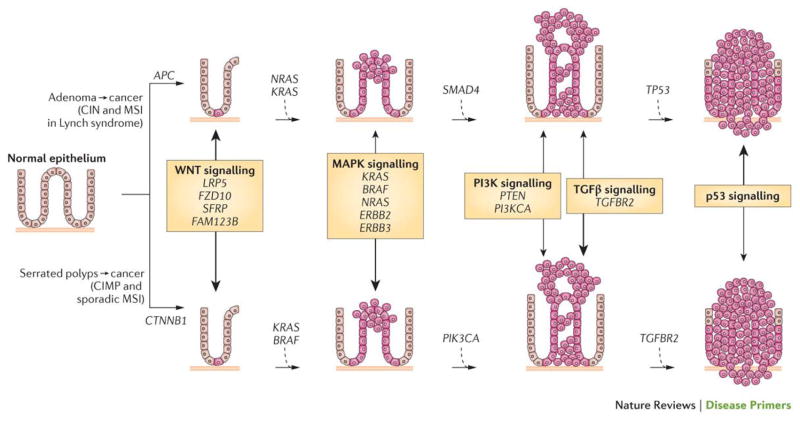
Figure 3. The polyp to colorectal cancer sequences.
Currently, two discrete normal colon to colorectal cancer sequences have been identified. Both sequences involve the progression of normal colon epithelial cells to aberrant crypt foci, followed by early and advanced polyps with subsequent progression to early cancer and then advanced cancer. The ‘classic’ or traditional pathway (top) involves the development of tubular adenomas that can progress to adenocarcinomas. An alternate pathway (bottom) involves serrated polyps and their progression to serrated colorectal cancer has been described in the last 5–10 years. The genes mutated or epigenetically altered are indicated in each sequence; some genes are shared between the two pathways whereas others are unique (for example, BRAF mutations and CpG Island Methylator Phenotype (CIMP) only occur in the serrated pathway). The signalling pathways deregulated during the progression sequence are also shown, with the width of the arrow reflecting the significance of the signalling pathway in tumour formation.
APC, adenomatous polyposis coli; CIN, chromosomal instability; CTNNB1, catenin-β1; FAM123B, family with sequence similarity 123B (also known as AMER1); FZD10, frizzled class receptor 10; LRP5, low-density lipoprotein receptor-related protein 5; MAPK, mitogen-activated protein kinase; MSI, microsatellite instability; PI3K, phosphatidylinositol 3-kinase; PI3KCA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit-α; PTEN, phosphatase and tensin homologue; SFRP, secreted frizzled-related protein; SMAD4, SMAD family member 4; TGFβ, transforming growth factor-β; TGFBR2, TGFβ _receptor 2.
Figure adapted from 229, Nature Publishing Group.