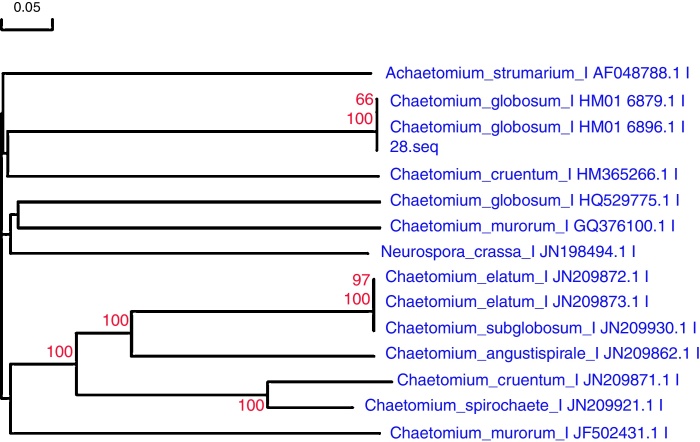
Fig. 2.
Cluster analysis of strain 28. The sequences that showed E = 0.0 and the highest % similarity with the amplified sequences were included for alignment and bootstrapping using CLUSTALX. This figure shows the phylogenetic relations of this isolate with related fungi. Thus, this strain was identified as Chaetomium globosum of Chaetomium sp. through the rDNA sequencing of ITS region data. This tree was constructed on the basis of the rDNA sequence (ITS1, 5.8S and ITS 2) by Neighbor-joining method. Bootstrap values >50% (1000 replicates) are shown at the branches. The bar indicates a 5% sequence divergence.