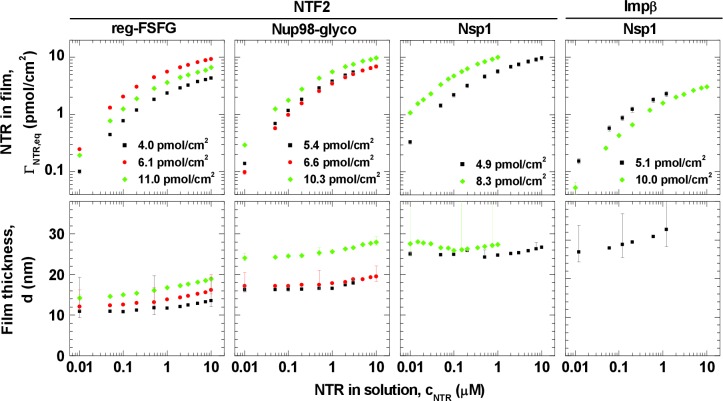
Figure 1. Isotherms of NTR binding (top row, log-log presentation) and FG domain film thickness evolution (bottom row, lin-log presentation) for NTF2 and Impβ binding to different FG domains (see labels at top) at selected FG domain grafting densities (visualized by distinct symbols and colors).
Error bars are shown for all data points in the binding isotherms, and for three selected data points (indicating the trends) per curve in the thickness isotherms. The data for Impβ binding to the 10.0 pmol/cm2 Nsp1 film were reproduced from Eisele et al. (2010); this data was acquired with Nsp1 carrying a His tag at the opposite end (N-terminus) compared to the other Nsp1 data in this study, in a separate SE measurement and no simultaneously recorded thickness data are available. Full experimental details are available in ‘Materials and methods’ and Figure 1—figure supplements 1–5; tabulated results are available in Figure 1—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.14119.005
Figure 1—figure supplement 1. Quality of purified recombinant proteins used in this study.
Figure 1—figure supplement 2. FG domains are anchored specifically and stably through their terminal His tag.
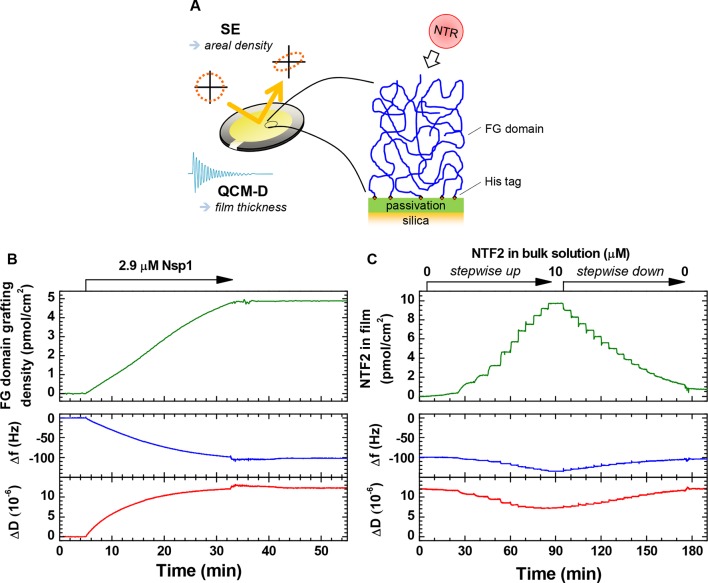
Figure 1—figure supplement 3. Schematic illustration of the experimental approach and representative data.
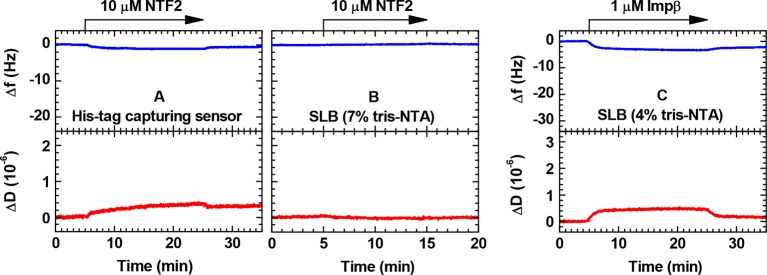
Figure 1—figure supplement 4. Controls for the binding of NTRs to His tag capturing surfaces monitored by QCM-D.
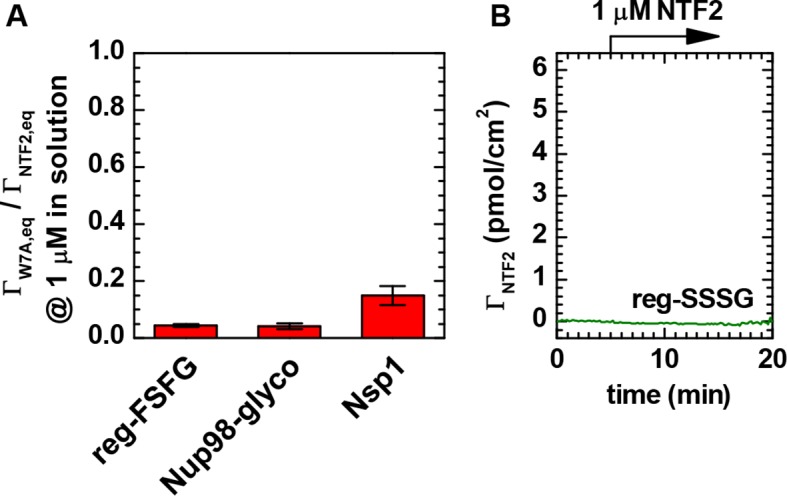
Figure 1—figure supplement 5. NTF2 binds all FG domains predominantly through its primary binding site.