Figure 4.
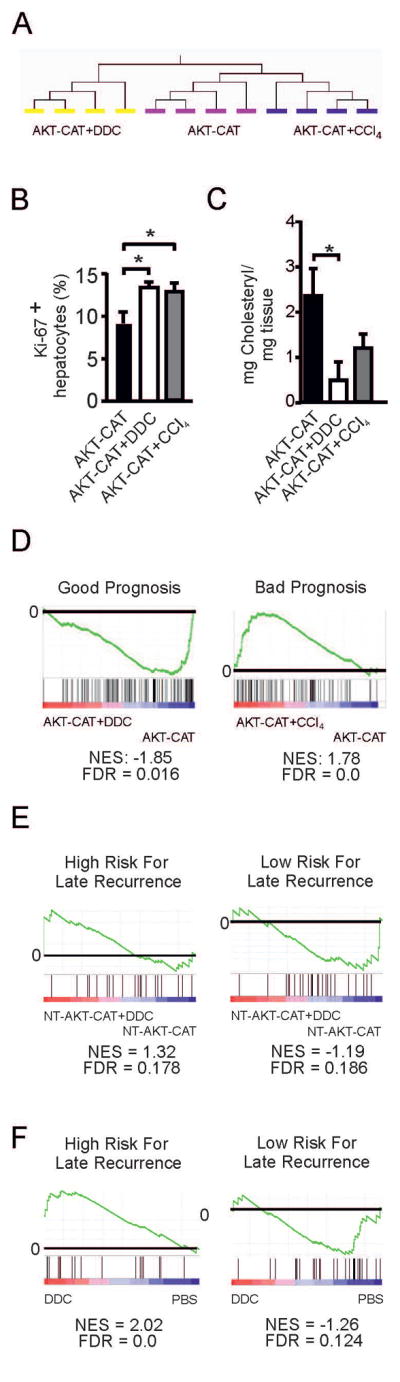
Transcriptome of AKT-CAT tumors generated in the background of chronic liver inflammation. A) Unsupervised hierarchical clustering of gene expression profiles from AKT-CAT, AKT-CAT+DDC and AKT-CAT+CCl4 tumors. B) Percentage of proliferating Ki-67 positive hepatocytes within total hepatocytes, measured by immunohistochemistry. Mean and SEM from 7 – 8 tumors/group. C) Cholesteryl ester concentration in AKT-CAT and AKT-CAT+DDC tumors. Mean and SEM of 4 tumor samples/group. D) GSEA of AKT-CAT and AKT-CAT+DDC tumors using a gene set with good prognosis (left) and of AKT-CAT and AKT-CAT+CCl4 tumors using a gene set with bad prognosis (right). E, F) GSEA of non-tumoral tissue of AKT-CAT and AKT-CAT+DDC mice (E) or mice treated with DDC only (F) using gene sets with low and high risk of late recurrence.
