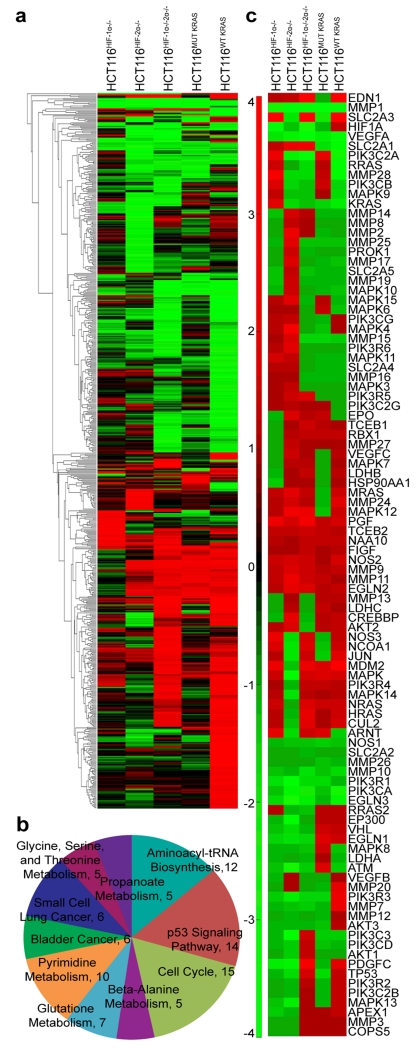
Figure 1.
HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells demonstrate similar transcript profiles. (a) Global gene expression affected by oncogenic KRAS showed significant overlap with genes affected by both HIF-1α and HIF-2α (parental HCT116 versus HCT116WT KRAS in comparison to parental HCT116 versus HCT116HIF-1α-/-HIF-2α−/−). (b) GO analysis revealed top molecular functions associated with HCT116HIF-1α-/-HIF-2α−/−. (c) Similar to effects on global gene expression, gene perturbations specific to the HIF pathway are more similarly affected in the HCT116HIF-1α-/-HIF-2α−/− and HCT116WT KRAS cells; compared to HCT116HIF-1α−/−, HCT116HIF-2α−/−, or HCT116MUT KRAS cells.