Figure 2. Regulation by covalent chromatin modifications and noncoding transcription.
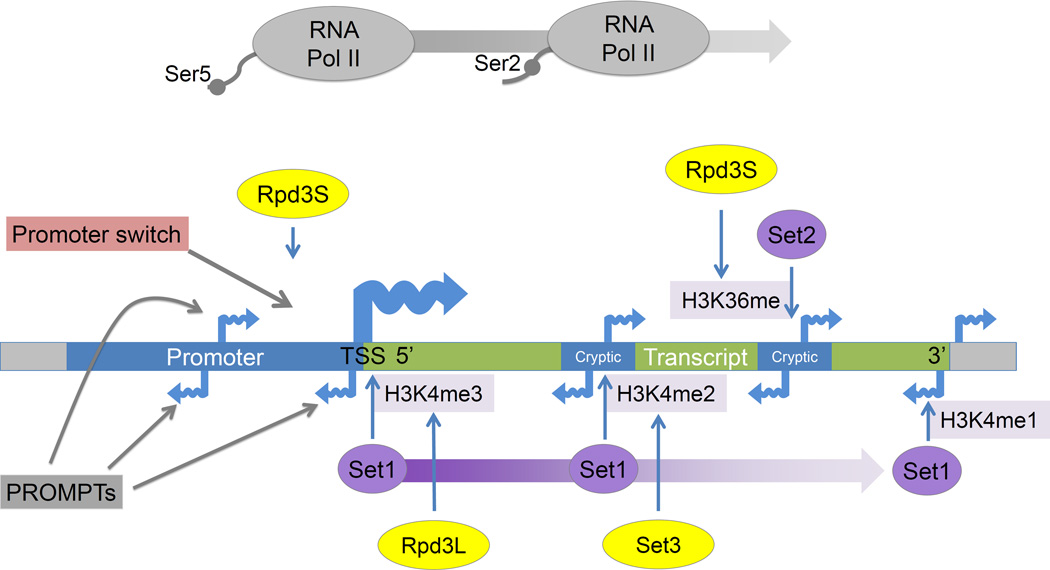
The interplay between histone methyltransferase (purple) and histone deacetylase (yellow) activities influences noncoding transcription. During transcription RNA Pol II is phosphorylated on its C terminal domain (CTD) first at serine 5 (Ser5P) near the 5' end of genes and then at serine 2 (Ser2P), allowing RNA Pol II to enter into the elongation phase of transcription, and establishing correct chromatin structure across transcribed regions [89]. Set1 interacts with RNA Pol II Ser5P and creates a gradient of H3K4 methylation starting with tri-methylation at the 5’ end and ending with mono-methylation at the 3’ end. These methyl marks are then targeted by two separate histone deacetylases. Rpd3L (the larger complex) deacetylates histones with H3K4me3 marks at the 5’ ends of transcripts [90]. Set3 deacetylates histones with H3K4me2 marks within gene bodies and represses 5’ proximal cryptic transcripts [91]. Set2 mediated H3K36 methylation takes place within gene bodies and towards the 3’ regions of the genes. This H3K36 modification is then targeted by Rpd3S (small complex) which removes the acetylation that co-occurs with transcriptional elongation, thereby suppressing cryptic transcripts originating within gene bodies near the 3’ end [92]. These transcripts are thought to arise because of chromatin remodeling concomitant with transcription, leading to increased accessibility of intragenic regions to TFs and Pol II [93]. Rpd3S has also been implicated in the repression of antisense transcripts, thereby promoting sense strand directed transcription [60].