Fig. 1.
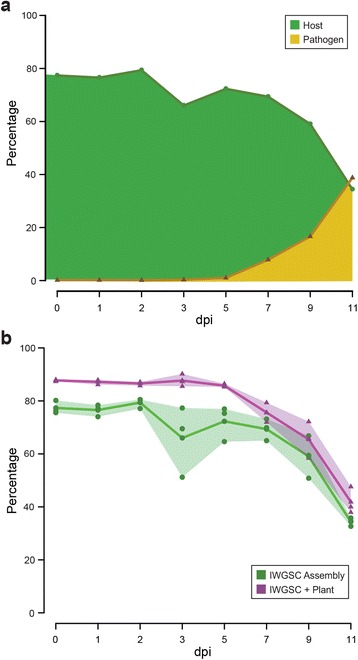
An initial depression in the percentage of reads mapping to the wheat genome early in infection could be restored by supplementing the wheat genome with plant-derived de novo assembled transcripts. a Alignment of RNA-seq data from the various time points during infection to both the wheat host and PST-130 pathogen reference genomes revealed a notable drop in the percentage of reads mapping to the wheat reference genome specifically at 3 days post inoculation (dpi). b The wheat reference genome, generated by the International Wheat Genome Sequencing Consortium (IWGSC), was supplemented with plant transcripts from a de novo assembly of the unmapped RNA-seq reads. Alignment of the RNA-seq data to this combined reference (“IWGSC + Plant”) restored the previous depression at 3 dpi
