Fig. 5.
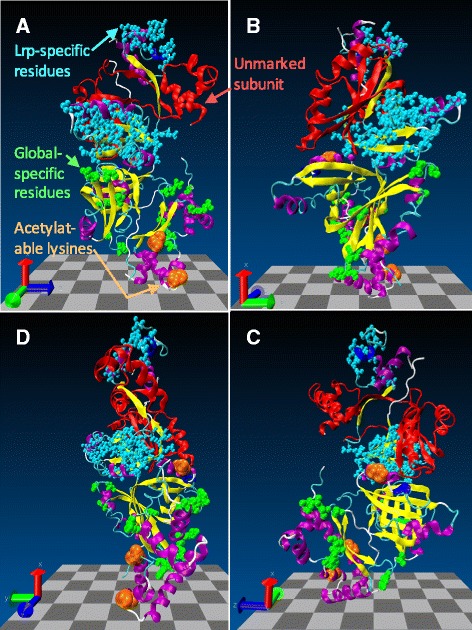
Visualization of residues of interest in context of Lrp 3D structure. The program VMD 1.9.2 was used to visualize half of an octameric ring of E. coli Lrp subunits (from PDB 2GQQ). VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. a-d are successive 90° rotations about the vertical axis. The topmost subunit has cyan spheres highlighting residues associated with Lrp-specific signatures (see Fig. 2c), the next subunit is shown in red without additional highlighting, the next subunit shows in orange spheres the lysines that can be acetylated (see Fig. 3a), and the bottom subunit shows in green spheres the residues associated with globally-acting Lrp orthologs (see Additional file 1: Table S1 for position numbers of all highlighted residues)
