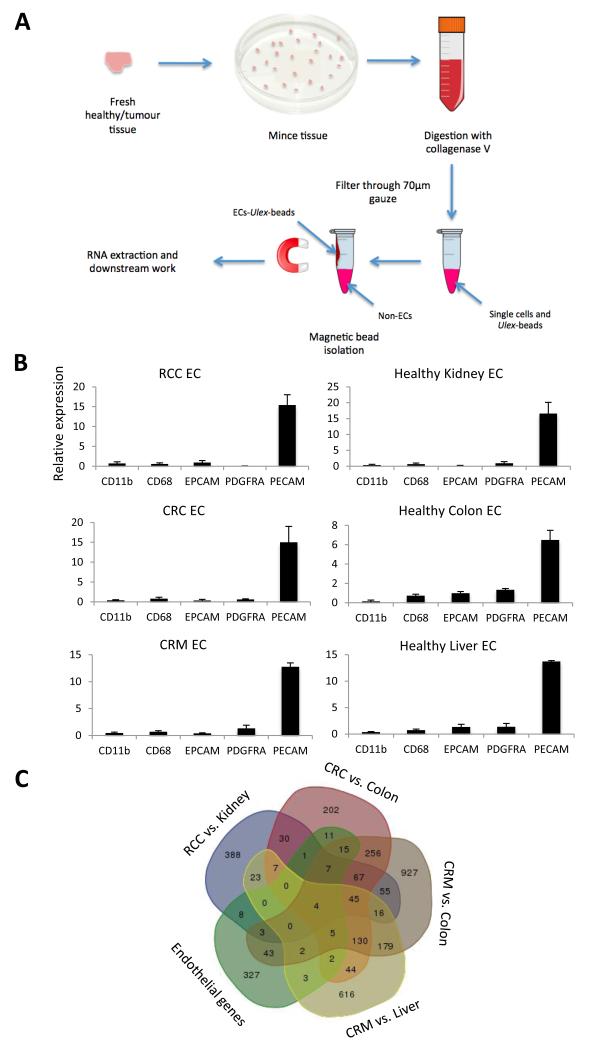
Figure 1.
Isolation of endothelial cells using Ulex lectin coated magnetic beads, for microarray analysis. A, the workflow of the main steps involved in the endothelial isolation procedure. B, confirmation of endothelial isolation efficiency by RTqPCR for markers of leukocytes (CD11b), macrophages (CD68), epithelium (EPCAM), smooth muscle (PDGFRA) and endothelium (PECAM) in the endothelial isolates (EC) from renal cell carcinoma (RCC, n = 8), colorectal carcinoma (CRC, n = 8), colorectal liver metastases (CRM, n = 7) and associated healthy tissues (n = 8,8,7), standardised to flotillin 2 (a house keeping gene) and normalised for marker expression in patient matched bulk tissue. The fold change of marker expression between the endothelial and bulk fraction is shown. Confidence limits are ± standard error of the mean (SEM). C, Venn diagram showing the commonality of genes at least 2 fold upregulated in 4 tumour vs. healthy tissue analyses and present in a list of known endothelial restricted genes.