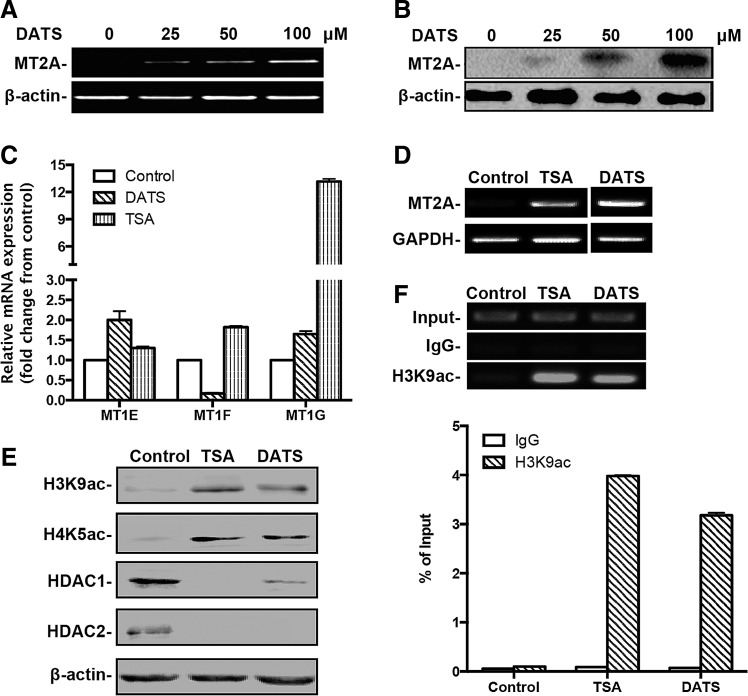
FIG. 4.
Epigenetic regulation of MT2A in GC cells by DATS. (A) RT-RCR and (B) Western blotting of MT2A expression in BGC823 cells after 12 h of treatment with DATS at different concentrations. (C) qPCR of the expression of MT1E, MT1F, and MT1G in BGC823 cells after DATS treatment (40 μM, 12 h). The mRNA of each gene was compared with its own control after normalization against GAPDH, presented as the mean ± standard deviation. (D) RT-PCR and (E) Western blotting of BGC823 cells after DATS (40 μM, 12 h) or TSA (5 μM, 24 h) treatment. (F) ChIP was performed on BGC823 cells after DATS (40 μM, 12 h) or TSA (5 μM, 24 h) treatment using antibody against H3K9ac or control IgG. Precipitated ChIP DNA fractions were analyzed by PCR using primers specific for the MT2A gene (<+1 kb from TSS). Top panel: representative conventional PCR products visualized on 1.5% agarose gel to analyze the association of H3K9ac to the MT2A gene. Bottom panel: qPCR analysis of the enrichment of H3K9ac at the MT2A gene, expressed as the percentage of input. Results are representative from at least three independent experiments. ChIP, chromatin immunoprecipitation; H3K9ac, histone 3 acetylation at lysine 9; TSA, trichostatin A; TSS, transcription start site.