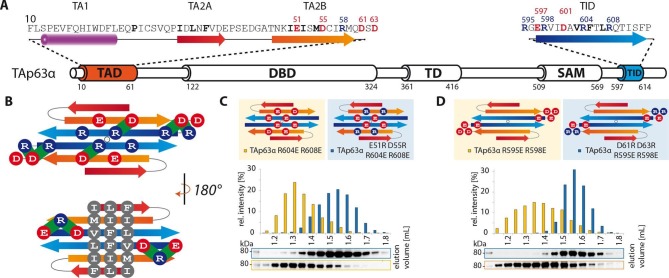
(A) Secondary structure prediction of TAD and TID. TA2B and
TID are predicted to form β-strands. (B) On validation,
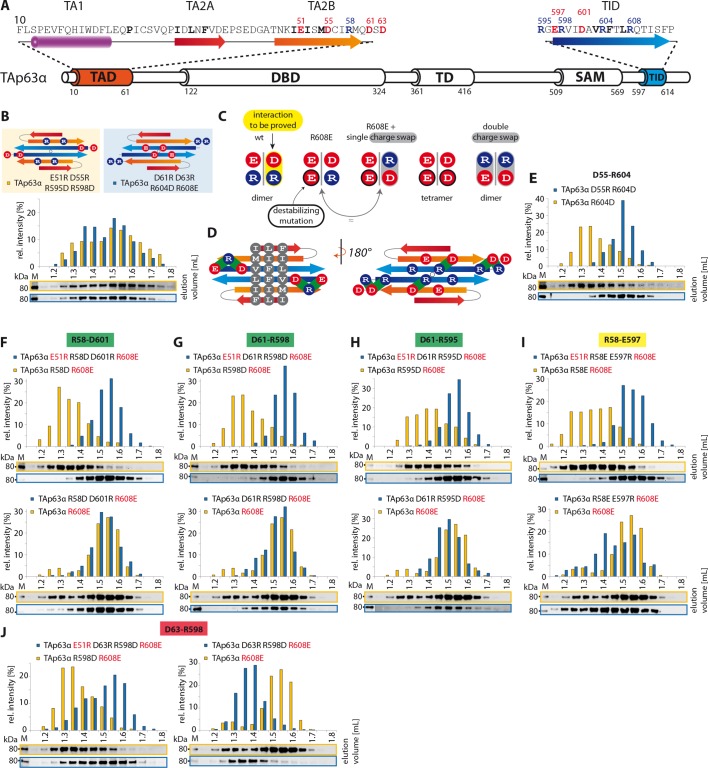
charges were swapped between presumably distant amino acids. SEC profiles
of TAp63α E51R D55R R595D R598D (orange) and TAp63α D61R D63R R604D R608E
(blue). (C) Charge swaps are used to reveal interactions
between charged amino acids across the presumed β-sheet formed by TA2B
and TID. A destabilizing mutation (arginine to aspartate or glutamate) is
introduced into TAp63α. Solely mutation R604D/E leads to the formation of
tetramers. Any other arginine in TA2B or TID mutated to aspartate or
glutamate does not change the oligomeric state. In order to break the
interaction between TA2B and TID, a second destabilizing mutation is
introduced that disrupts the dimeric state and leads to the formation of
tetramers. Additional compensating mutations in the double charge swap
recover the dimeric state. To prove a single interaction between two
differently charged amino acids, they are swapped in presence of a
destabilizing mutation (R608E) resulting in a triple mutant. Similar SEC
profiles of the destabilizing and the triple mutation prove the
interaction between the swapped amino acids. (D) Extensive
charge swap experiments (shown in E-I) revealed interactions
(shown in green) between TA2B and TID. (E) SEC profiles of a
single charge swap between R604 and D55 (blue) and the destabilizing
mutation R604D show a direct interaction between D55 and R604.
(F,G,H,I,J)
SEC profiles of double arginine mutants (top, orange) and double charge
swaps (top, blue). SEC profiles of triple mutants (bottom, blue) and the
R608E mutant (bottom, orange) should be identical to verify the
interaction shown in bold (top). For comparison identical western blots /
SEC profiles of TAp63α R608E are shown on bottom. (F) SEC
profiles of TAp63α R58D R608E (top, orange), charge swap TAp63α E51R R58D
D601R R608E (top, blue), triple mutant TAp63α R58D D601R R608E (bottom,
blue) and mutant TAp63α R608E (bottom, orange). (G) SEC
profiles of TAp63α R598D R608E (top, orange) and charge swap TAp63α E51R
D61R R598D R608E (top, blue), triple mutant TAp63α D61R R598D R608E
(bottom, blue) and mutant TAp63α R608E (bottom, orange). (H)
SEC profiles of TAp63α R595D R608E (top, orange) and charge swap TAp63α
E51R D61R R595D R608E (top, blue), triple mutant TAp63α D61R R595D R608E
(bottom, blue) and mutant TAp63α R608E (bottom, orange). (I)
SEC profiles of TAp63α R58E R608E (top, orange) and charge swap TAp63α
E51R R58E E597R R608E (top, blue), triple mutant TAp63α R58E E597R R608E
(bottom, blue) and mutant TAp63α R608E (bottom, orange). (J)
SEC profiles of TAp63α R598D R608E (left, orange, identical blot/profile
as shown in G) and charge swap TAp63α E51R D63R R598D R608E
(left, blue), triple mutant TAp63α D63R R598D R608E (right, blue) and
mutant TAp63α R608E (right, orange).