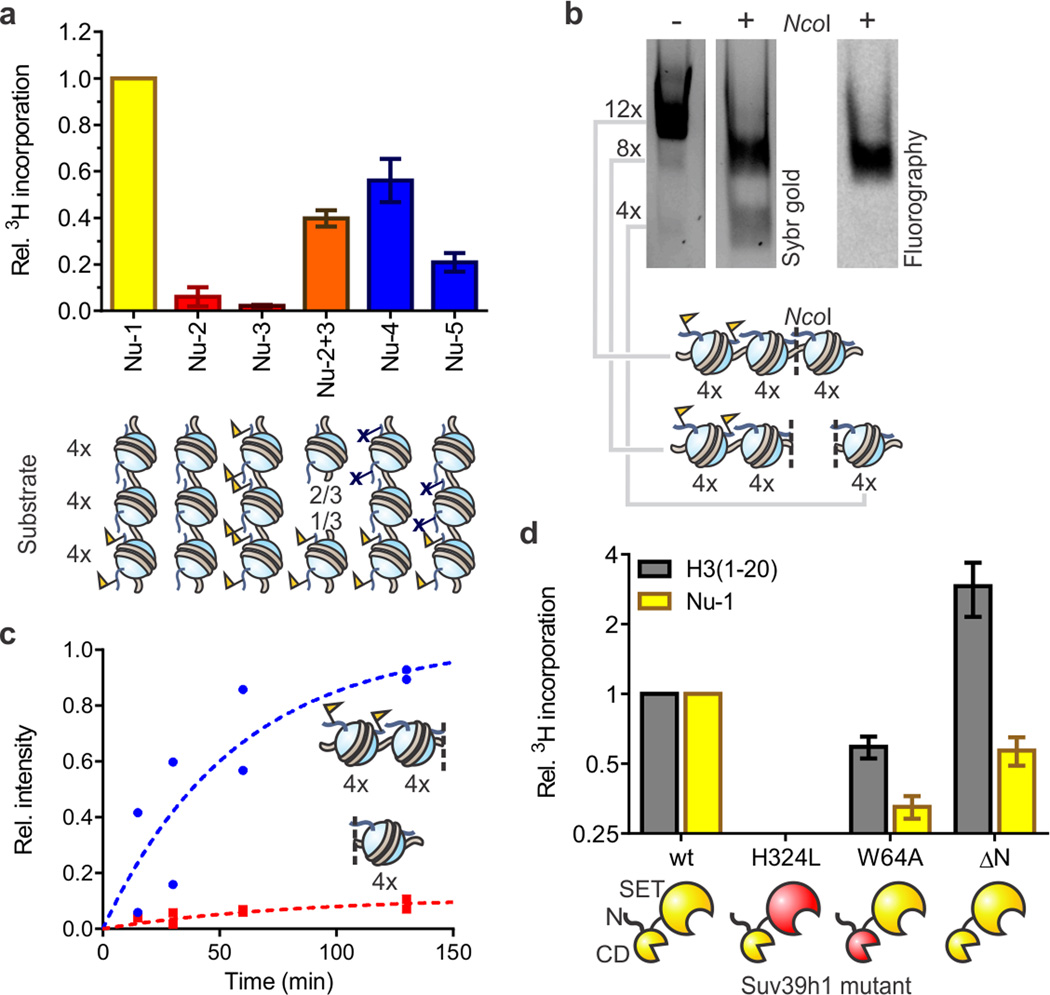
Figure 2. Reconstitution of Suv39h1-dependent heterochromatin spreading in vitro.
(a) Substrate preference of Suv39h1-catalyzed spreading. Arrays Nu-2 and Nu-3 represent homotypic templates (red) carrying either H3K9me0 or H3K9me3, respectively. Inter-fiber spreading is evaluated with a 2:1 mixture of arrays Nu-2 and Nu-3 (orange) to match the stoichiometry of histones in array Nu-1 (yellow). Arrays Nu-4 and Nu-5 (blue) contain H3K9R mutations to block spreading. HMT activity was measured using 3H-SAM as the co-factor in the presence of 5 mM MgCl2. Scintillation counts are normalized to the values determined with heterotypic array Nu-1, without additional normalization for number of substrate sites. Error bars, s.e.m. (n = 3). (b) Suv39h1 preferentially methylates nucleosomes adjacent to preinstalled H3K9me3 marks. Arrays were methylated with 3H-SAM in the presence of 0.5 mM MgCl2 and subsequently digested with NcoI, separated by native gel electrophoresis and analyzed by Sybr gold staining (left) and fluorography (right). See Supplementary Fig. 8 for more details. (c) Time course of Suv39h1-dependent spreading of the H3K9me3 mark. Individual values, normalized to the sum of the values for the 4mer and 8mer obtained at 130 min, from two independent measurements are shown. (d) HMT activity of Suv39h1 variants with an H3 peptide encompassing residues 1–20 (gray) or array Nu-1 (yellow). Measurements are normalized to the values obtained with wild-type Suv39h1 and the respective substrate, and plotted on a log2 scale. Error bars, s.e.m. (n = 3).