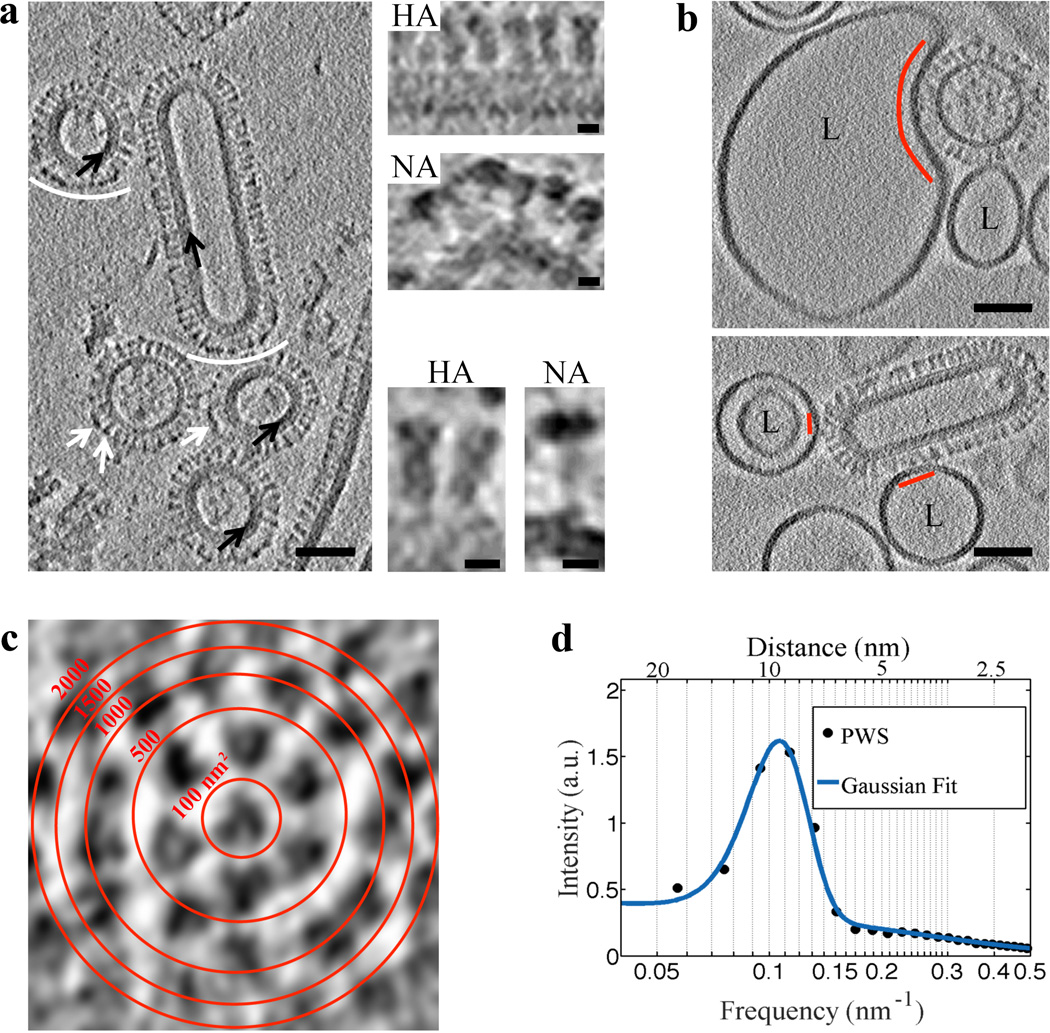
Figure 1. VLP structural features are resolved using VPP and indicate that influenza VLP carrying G1S mutated HA engage 1–7 HA glycoproteins on binding to liposomes.
a, A tomogram slice (3 nm thick) of WT VLP acquired using VPP and calculated by the weighted-back projection, capturing spherical particles without or with matrix layer (black arrows), and a filamentous particle. NA spikes (marked by white arrows or arcs) are characterized by a large globular domain and thin stalk as opposed to HA which has a cylindrical shape7. NA clusters (white arcs) observed in VLP are also found in filamentous influenza virus27. The tomographic slice in a is representative of VLP found in 7 tomograms collected on the same grid. Scale bars: 50 nm. Magnified views of HA and NA spikes. Scale bars: 5 nm. b, VLP carrying the G1S mutation in HA, when mixed with liposomes (labeled L) containing 16 mol % cholesterol and incubated for 20 minutes at neutral pH and subsequently subjected to pH 5 at 37°C for approximately 2 minutes demonstrate binding (binding area indicated by red arc and lines). VPP-cET tomogram slices (3 nm thick) calculated by the weighted-back projection method, showing VLP that are bound to liposomes but did not undergo conformational change. Scale bars: 50 nm. c, Tomogram slice of a G1S VLP surface showing transverse cross-sections of glycoproteins. Red concentric rings mark area boundaries within a 50×50 nm image. d, Radially averaged power spectrum of glycoprotein cross-sections shows an average 9.5±1.8 nm spacing (centers of the glycoproteins) on the surface of the G1S VLP. The tomographic slice b, is representative of 21 VLP-liposome binding events found in 12 tomograms collected on 2 independent grids as a technical replica of the sample.