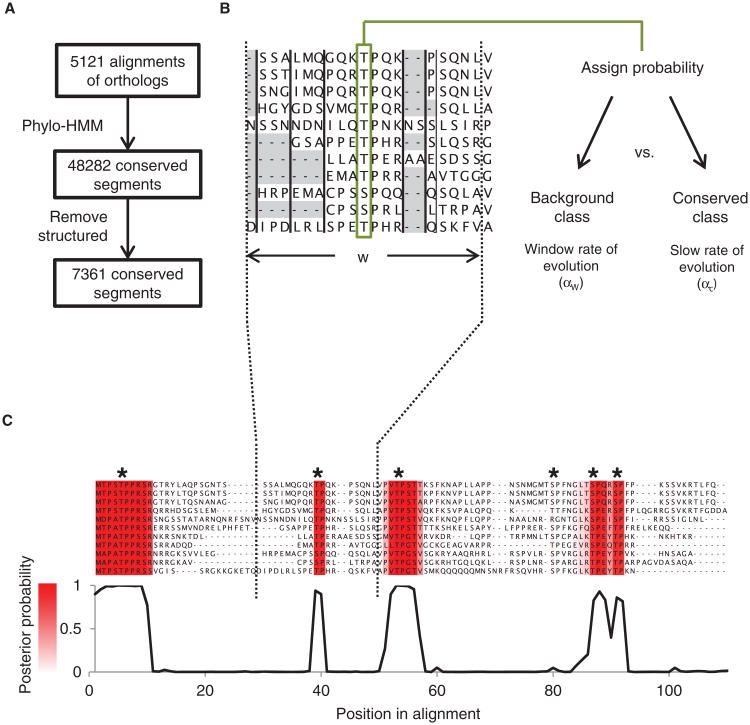
Fig. 1.
Schematic of the phylo-HMM approach. (A) Flowchart of the computational framework to detect conserved short linear motifs in disordered regions of multiple sequence alignments. (B) The rate of evolution for an alignment column (framed in green) is compared to a rate of evolution over a window (w) adjacent to the column. The probability that the column is within the preferentially conserved class is computed. The framework takes advantage of the amino acid substitutions inferred in columns of the alignment and the pattern of insertions and deletions (illustrated as gray highlights) in blocks of the multiple sequence alignment (separated by vertical black lines). (C) A posterior probability trace of the region 1 to 110 in the alignment of Sic1 (corresponding to amino acid positions 1 to 100 in S. cerevisiae). Four strongly conserved segments are detected by the phylo-HMM approach and these overlap with experimentally reported phosphorylation sites in Sic1 (indicated by stars), which are required for Cdc4 binding. The intensity of the red color represents the posterior probability of the conserved state.