Figure 2.
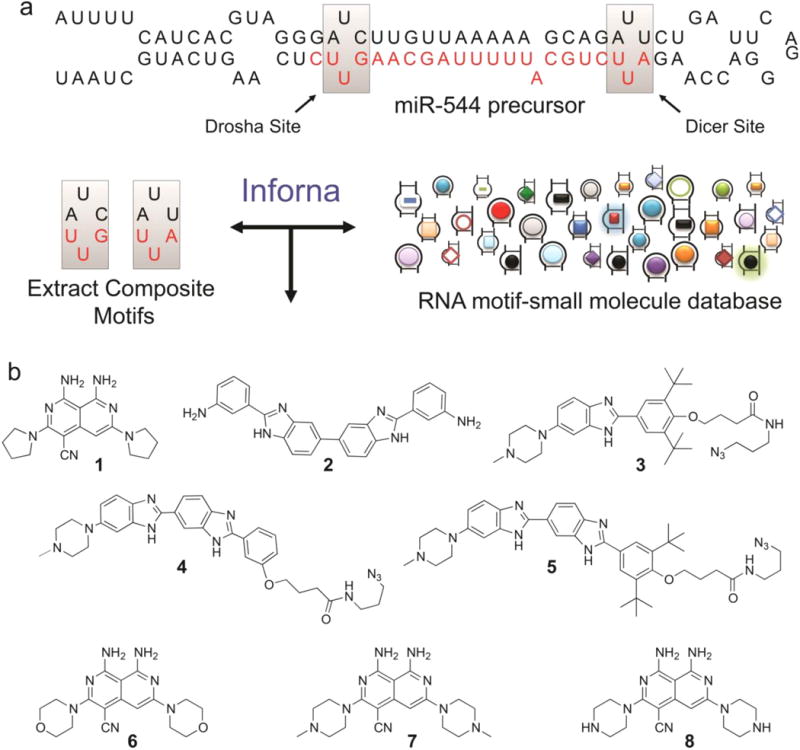
Identification of small molecules that impede miR-544 biogenesis via direct binding. (a) Hairpin structure of miR-544 precursor showing 1 × 1 nucleotide UU internal loops located in the Drosha and Dicer processing sites targeted by 1 and 2. (b) Compounds (1–5) identified from similarity searches between miR-544 precursor motifs and a fully characterized set of RNA motif small molecule interactions. Three scaffold modified derivatives of compound 1 (6–8) in which the pyrrolidine is substituted with morpholine, methylpiperazine, or piperazine.
