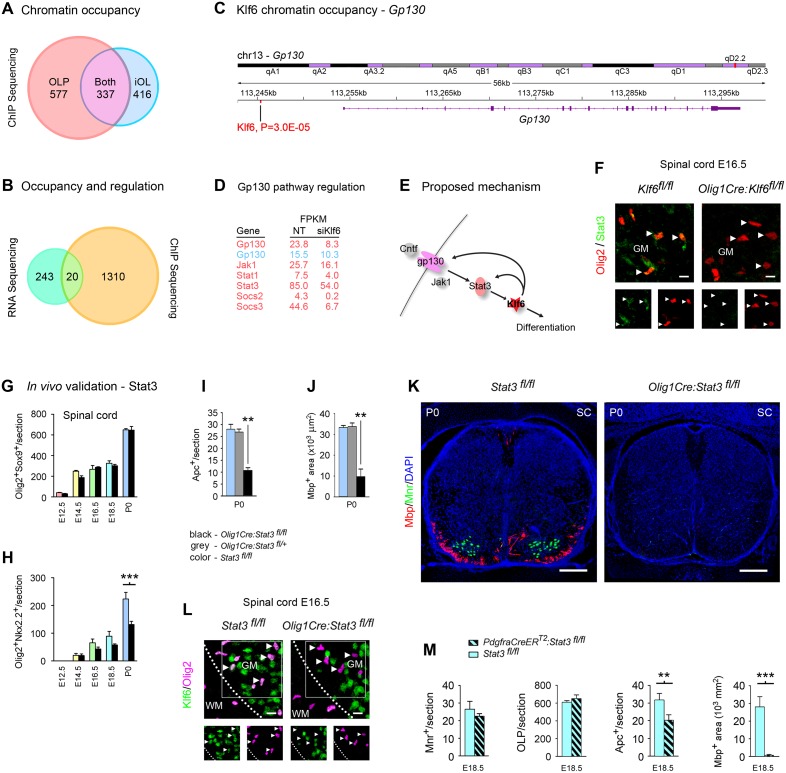
Fig 5. Stat3 inactivation in vivo produces failure of myelination, similar to Klf6 inactivation.
(A) Graphical overview of genome-scale chromatin occupancy (ChIP-seq) data for Klf6 in primary mouse OLP and iOL. ChIP-seq analysis identifies 577 peaks of Klf6 chromatin binding within 20 kb of transcription start sites in OLP, 416 in iOL, and 337 that are shared. See also S4 Table. (B) Cross-referencing this ChIP-seq dataset to RNA-seq data in S1 and S2 Tables identifies 20 genes as prospective directly-regulated Klf6 effectors. This cohort is presented in S5 Table. (C,D) ChIP-seq identifies Klf6 binding to the promoter of the gp130 gene (C), which is also amongst multiple gp130-Stat3 signaling pathway components identified by RNA-seq as Klf6-regulated (D, and see S1–S3 Tables). These findings suggest that Klf6 exerts feedback upon the gp130-Stat3 pathway that drives its expression (E). (F) Confocal images of spinal cords of E16.5 Olig1Cre:Klf6fl/fl and Klf6fl/fl control embryos labeled for Stat3 and Olig2. Areas illustrated are from grey matter (GM). Levels of Stat3 are reduced in the oligodendrocyte lineage in Olig1Cre:Klf6fl/fl samples. Arrowheads mark representative cells, which are shown at higher magnification below. See also S6A Fig. (G–M) Confocal analysis of spinal cords from mice with Stat3 inactivation targeted to OLP (Olig1Cre:Stat3fl/fl and PdgfracreERTM:Stat3fl/fl). The oligodendrocyte phenotype of these mice resembles that produced by conditional Klf6 inactivation (see Fig 2). At E12.5–14.5, OLP numbers in conditional Stat3 mutants are comparable to Stat3fl/f controls, and early events in differentiation occur normally (G,H). However, selective loss of differentiating cells is seen from E16.5 onward (H,I), and myelination is profoundly disrupted (J,K, see also S6B and S6C Fig). Stat3 inactivation produced by Olig1Cre also results in loss of motor neurons, which share the same origin in the pMN domain (K, and see S6D Fig). (L) Spinal cords of E16.5 Olig1Cre:Stat3fl/fl and control Stat3fl/fl embryos, labeled for Klf6 and Olig2. The border between grey (GM) and white matter (WM) is marked, and representative cells are arrowed. Conditional Stat3 inactivation produces defective Klf6 expression. (M) Conditional Stat3 inactivation in OLP using PdgfracreERTM results in selective failure of OLP differentiation and myelination. Motor neurons are unaffected. Data are mean ± SEM. Statistics, (G–J) ANOVA plus Bonferroni test, (M) Student’s t test, **p < 0.01, ***p < 0.001. Scalebars, (F,L) 5 μm, (K) 250 μm. Data are representative of two to six mice per genotype per time point. Results in (F–M) are from lumbar sections. ChIP-seq data are presented in full in S4 Table, RNA-seq and ChIP-seq intersect data are presented in full in S5 Table, and both datasets are available on the GEO website (http://www.ncbi.nlm.nih.gov/geo/) (Accession number GSE79245). Individual values for all other quantifications are in S1 Data.