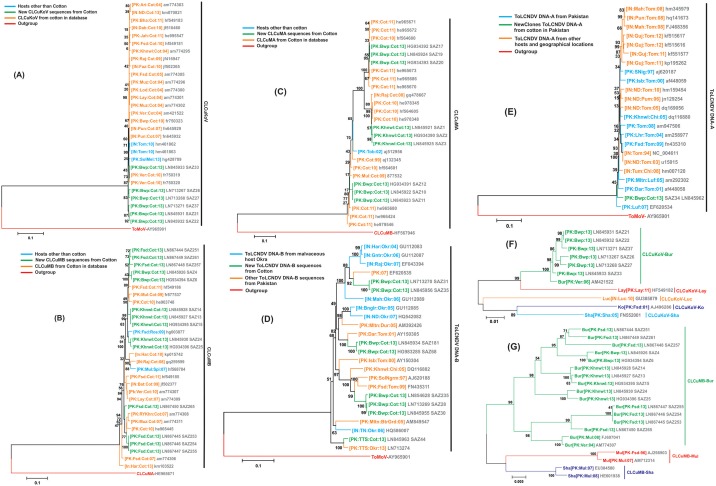
Fig 2. Phylogenetic analysis of the sequences of begomoviruses, and satellites obtained in the study.
Trees were constructed from alignments of the sequences of (A and F) Cotton leaf curl Kokhran virus (CLCuKoV), (B and G) Cotton leaf curl Multan betasatellite (CLCuMB), (C) Cotton leaf curl Multan alphasatellite (CLCuMA), (D) Tomato leaf curl New Delhi virus (ToLCNDV) DNA-B and (E) ToLCNDV DNA-A using the Neighbor-Joining method. Isolates in green were obtained in the study described here. The numbers at nodes represent percentage bootstrap scores (1000 replicates). The strain descriptors (in square brackets) in each case give country, location, host and year of sampling. For each isolate the database accession number is given. The trees were arbitrarily rooted on the sequence of Tomato mottle virus (ToMoV) (A, D and E), CLCuMA (B) and CLCuMB (C) as outgroup. The strains of CLCuKoV given in panel F are Burewala (Bur), Layyah (Lay), Lucknow (Luc), Kokhran (Kok) and Shadadpur (Sha). The strains of CLCuMB given in panel G are given as Burewala (Bur), Multan (Mul) and Shadadpur (Sha).