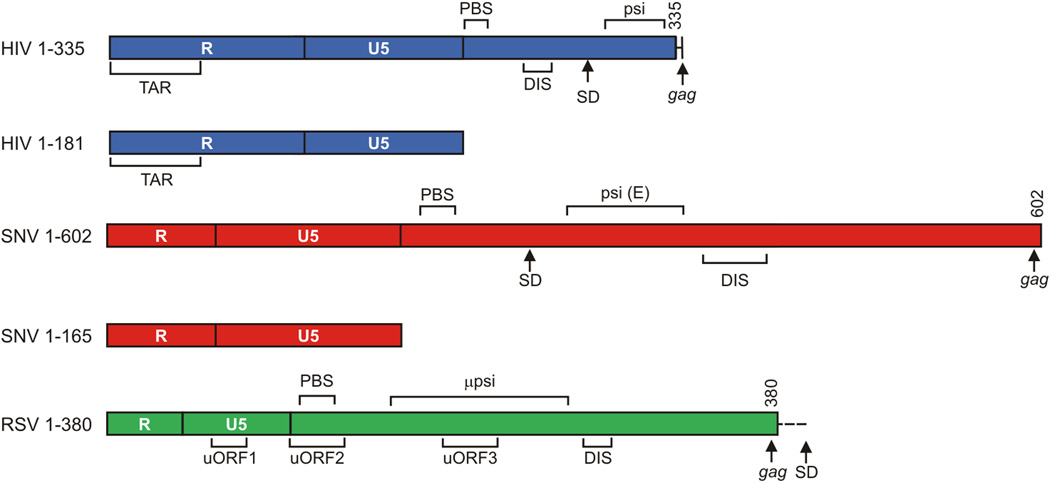
Figure 1.
A: Drawing of the retroviral transcripts used in this study. Numbers indicate nucleotide position relative to the start of the viral RNA. TAR: trans-activation response element; PolyA: Polyadenylation Signal; PBS: Primer Binding Site; 5’ss: 5’ Splice Site; DIS: Dimerization initiation signal; uORF: Upstream Open Reading Frame. µpsi coordinates from pATV8.
B: Immunoblot analysis of candidate 5’ UTR binding proteins relative to negative controls. Evaluation of stringency achieved in the RNA affinity chromatography of Hela cell extract. RNA affinity chromatography was performed on the indicated RNAs and eluates were collected after progressive wash cycles and equivalent volumes were subjected to SDS-PAGE and Western blot (WB) with the indicated antiserum. HeLa Input demonstrates abundant DHX30, YBX1 and GAPDH in the lysate and chemiluminscence indicates DHX30 and YBX1 are enriched relative to MYC 5’ UTR or RNA-free resin (Beads Only) and GAPDH.