FIGURE 6.
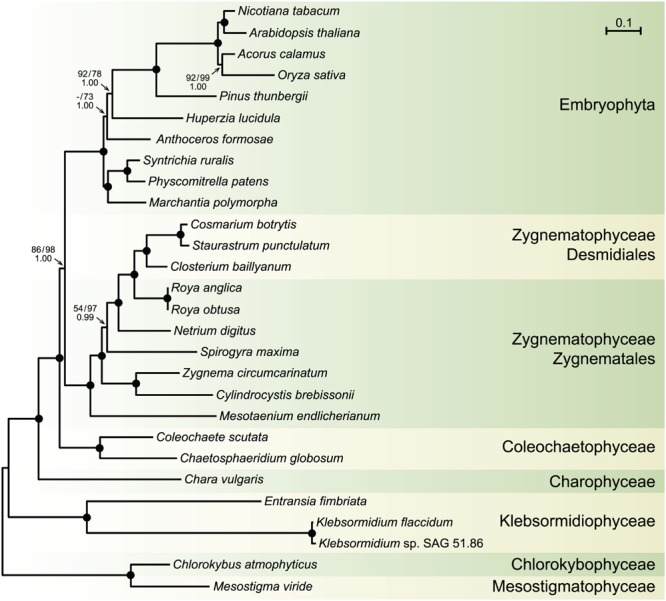
Phylogeny of streptophytes inferred using amino acid and nucleotide data sets assembled from 88 chloroplast protein-coding genes. The best-scoring tree inferred with Phylobayes from the PCG-AA data set under the CATGTR + Γ4 model is presented. Mesostigma and Chlorokybus were used as outgroup. Bootstrap support (BS) and posterior probability (PP) values are reported on the nodes: on top from left to right, are shown the BS values for the RAxML analyses of the PCG-AA and PCG12 data sets, respectively; the PP values of the Phylobayes CATGTR + Γ4 analysis are presented under the BS values. A black dot indicates that the corresponding branch received maximum support in all three analyses; a dash represents a BS value <50%. The scale bar denotes the estimated number of amino acid substitutions per site. The GenBank accession numbers of the streptophyte algal chloroplast genomes included in the PCG-AA and PCG12 data sets are provided in Table 1. The GenBank accession numbers of the land plant chloroplast genomes are as follows: Marchantia polymorpha NC_001319, Physcomitrella patens NC_005087, Syntrichia ruralis NC_012052, Anthoceros formosae NC_004543, Huperzia lucidula NC_006861, Pinus thunbergii NC_001631, Oryza sativa NC_001320, Acorus calamus NC_007407, Arabidopsis thaliana NC_000932, Nicotiana tabacum NC_001879.
