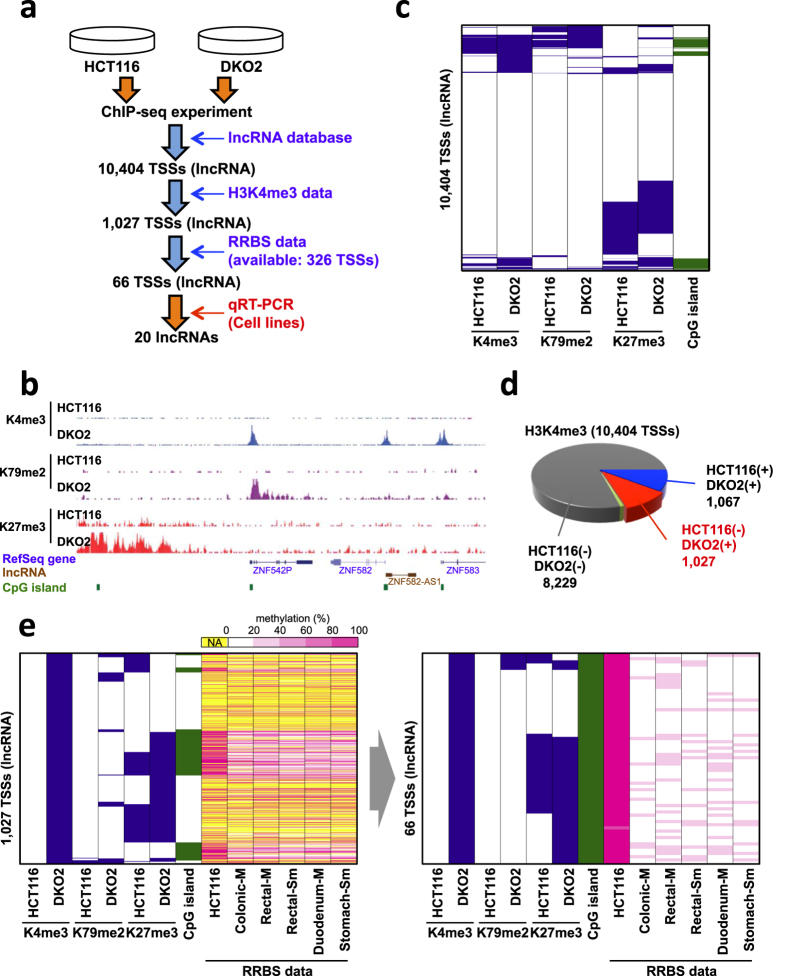
Figure 1. Screen for epigenetically silenced lncRNA genes in CRC.
(a) Workflow of the screen to identify lncRNAs silenced in association with aberrant CGI methylation in CRC. (b) Representative results of a ChIP-seq analysis in HCT116 and DKO2 cells. (c) Heat map showing the presence (blue) or absence (white) of histone modifications (H4K4me3, H3K79me2 and H3K27me3) at the TSS regions of lncRNA genes in HCT116 and DKO2 cells. The presence (green) or absence (white) of a CGI is also indicated on the right. (d) The fraction of TSSs with an H3K4me3 mark in HCT116 and DKO2 cells. Shown are the numbers of TSSs with the indicated H3K4me3 status in the two cell lines. (e) Heat maps showing the histone modifications at selected TSSs in CRC cells. Shown is the DNA methylation status obtained from RRBS data sets for HCT116 and normal gastrointestinal tissues. A set of 1,027 TSSs with increased H3K4me3 in DKO2 cells is shown on the left, and 66 TSSs with cancer-specific CGI methylation are shown on the right. Colonic-M, colonic mucosa; Rectal-M, rectal mucosa; Rectal-Sm, rectal smooth muscle; Duodenum-M, duodenum mucosa; Stomach-Sm, stomach smooth muscle; NA, not available.