Fig. 1.
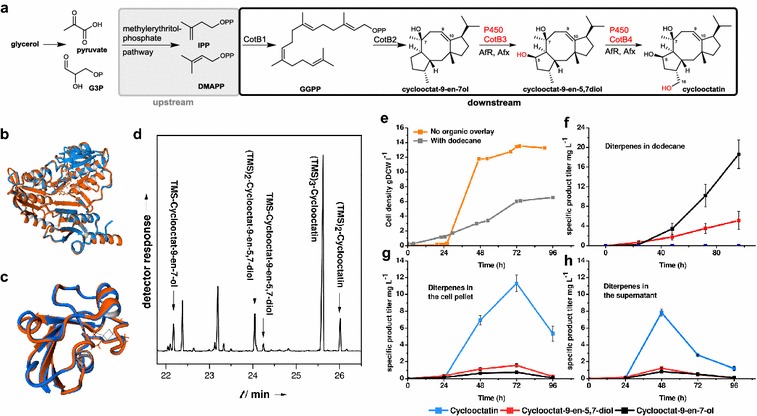
Native and heterologous reconstituted cycloocatin biosynthesis with non-native redox partners. a The cyclooctatin biosynthetic pathway encodes a GGDP synthase (CotB1), a diterpene cyclase (CotB2) and two P450 type hydroxylases (CotB3, CotB4). The structure alignments of AfR (orange) with PdR (blue) (b) and Afx (orange) with Pdx (blue) (c) are shown. Despite the low primary sequence similarities structure alignments of PdR with AfR as well as Pdx with Afx have both a root mean square deviation (RMSD) of 0.1 Å over 404 and 106 aligned amino acid residues, respectively. d GC–MS chromatogram of a silylated cellular extract from the cyclooctatin producing E. coli strain harvested after 48 h of growth. e Monitored cell density during fermentation is displayed (±dodecane). f Production of cyclooctatin and its biosynthetic intermediates in the dodecane organic phase is shown. g, h Monitored product distribution during fermentation in absence of dodecane
