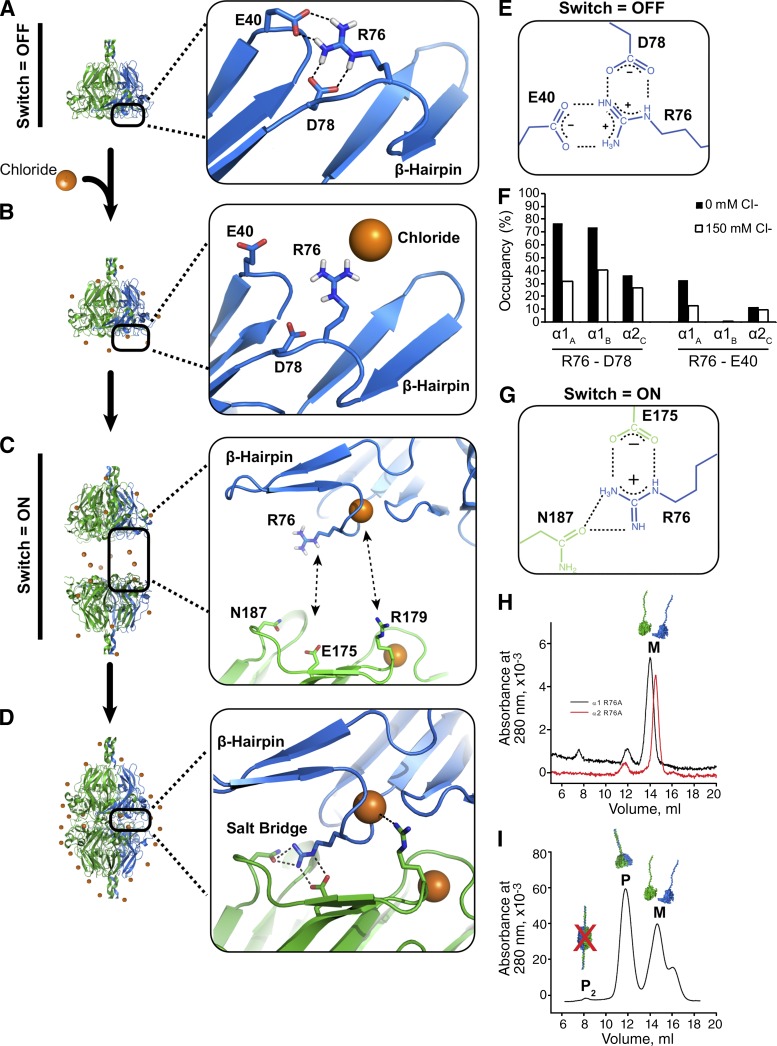
Figure 5.
Cl− triggers a molecular switch enabling network assembly. (A) Without Cl−, R76 forms intramolecular salt bridge with D78 and/or E40 in MD simulations. (B) Extracellular Cl− disrupts R76-D78 salt bridge by electrostatic screening. (C) Specific binding activity of Cl− causes the ion to coordinate the R76 backbone amide of R76, thus orienting the side toward an opposing NC1 timer. (D) Each bound Cl− ion can coordinate two distinct electrostatic interactions, up to 12 such interactions per hexamer, including six bridging-networked salt bridges. (E) Molecular structure of interactions among R76, E40, and D78. (F) Occupancy plot shows simulated hydrogen bond occupancies of R76 in 0 mM Cl− (closed bars) and 150 mM Cl− (open bars), indicating that Cl− disrupts R76 intramolecular interactions. (G) Molecular structure of electrostatic interactions that comprise the bridging-networked salt bridge. (H–I) R76A mutations prevent formation of hexamers. SEC profiles of purified α1-R76A (black line) and α2R76A (red line) r-monomers (H). SEC profile after mixing and incubation of both r-monomers in 100 mM NaCl showing the formation of protomers (P) but not protomer dimers (P2 and I).