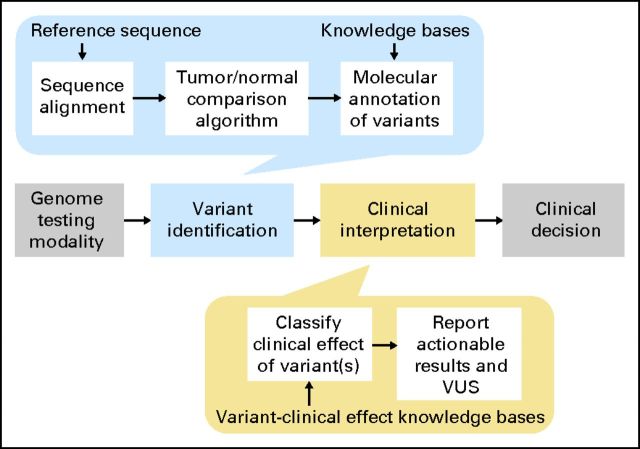
Fig 1.
Clinical workflow for tumor genome analysis and interpretation. The workflow begins with the genome-testing modality that may or may not have digital output. Variant identification may be done manually or with the assistance of automated algorithms, depending on the modality. Automated methods for variant identification include several processes shown at the top, including algorithms for sequence alignment that take as input a reference sequence, algorithms for comparison of the tumor and normal genome, and variant annotation. Clinical interpretation of the variant is a process that includes a determination of the size of variant effect and its interaction with other variants as well as an analysis of the strength of the evidence of the effect. These processes require the use of knowledge bases of variant–drug–disease relationships. Actionable results are reported as well as variants of unknown significance (VUS). The process culminates with a clinician using the information to make a clinical decision.