Abstract
Synthesis and regulation of catecholamine neurotransmitters in the central nervous system are implicated in the pathogenesis of a number of neuropsychiatric disorders. To identify factors that regulate the presynaptic synthesis of catecholamines, we tested the hypothesis that the rate-limiting enzyme of the catecholamine biosynthetic pathway, tyrosine hydroxylase (TH), is locally synthesized in axons and presynaptic nerve terminals of noradrenergic neurons. To isolate pure axonal mRNA and protein, rat superior cervical ganglion sympathetic neurons were cultured in compartmentalized Campenot chambers. qRT-PCR and RNA in situ hybridization analyses showed that TH mRNA is present in distal axons. Colocalization experiments with nerve terminal marker proteins suggested that both TH mRNA and protein localize in regions of the axon that resemble nerve terminals (i.e., synaptic boutons). Analysis of polysome-bound RNA showed that TH mRNA is present in polysomes isolated from distal axons. Metabolic labeling of axonally synthesized proteins labeled with the methionine analog, L-azidohomoalanine, showed that TH is locally synthesized in axons. Moreover, the local transfection and translation of exogenous TH mRNA into distal axons facilitated axonal dopamine synthesis. Finally, using chimeric td-Tomato-tagged constructs, we identified a sequence element within the TH 3′UTR that is required for the axonal localization of the reporter mRNA. Taken together, our results provide the first direct evidence that TH mRNA is trafficked to the axon and that the mRNA is locally translated. These findings raise the interesting possibility that the biosynthesis of the catecholamine neurotransmitters is locally regulated in the axon and/or presynaptic nerve terminal.
Keywords: catecholamine neurotransmitter, axonal mRNAs, axonal localization, intra-axonal translation, cis-acting regulatory element, superior cervical ganglion, dopamine
INTRODUCTION
Tyrosine hydroxylase (TH) catalyzes the rate-limiting step in the biosynthesis of the catecholamine neurotransmitters by converting tyrosine to dihydroxyphenylalanine (L-DOPA). According to present understanding, in catecholaminergic neurons, TH is synthesized exclusively in the cell body and is subsequently transported to its ultimate site of function, i.e., the axon and/or presynaptic nerve terminal (Jarrott and Geffen 1972; Axelrod 1974). However, studies have reported the presence of TH mRNA in the neocortex, striatum, and cerebellum, all brain regions that are devoid of catecholamine-synthesizing cells but receive catecholaminergic innervation (Lewis et al. 1991; Melia et al. 1994). In particular, in the cerebellum, levels of TH mRNA could be modulated by the administration of the catecholamine depleting agent, reserpine, suggesting that local synthesis of TH might function to facilitate the restoration of neurotransmitter levels (Melia et al. 1994). Together, these observations provided indirect evidence to suggest that TH mRNA may be trafficked to axons and nerve terminals of catecholaminergic neurons where it may be locally translated.
It is now well accepted that a relatively large and highly diverse population of axonal mRNAs are localized and translated in the axons and nerve terminals of neurons (for review, see Holt and Schuman 2013; Crispino et al. 2014; Scott et al. 2015). These axonal mRNAs code for several functional categories of proteins including cytoskeletal elements, ribosomal proteins, and translation factors, as well as metabolic enzymes and nuclear-encoded mitochondrial mRNAs (for review, see Kaplan et al. 2009). The targeting of specific mRNAs to the distal structural/functional domains of the neuron is driven by sequences located in the RNA itself (cis-acting elements), that are often found in the 3′ untranslated region (UTR) of the mRNA (Jambhekar and DeRisi 2007; Aschrafi et al. 2010; Doyle and Kiebler 2011; Meer et al. 2012).
These observations raise the possibility that, in addition to being synthesized in the cell body, TH could also be locally synthesized in the axon and nerve terminal. Local synthesis of TH would allow for the activity-dependent regulation of enzyme availability and would facilitate a rapid response to alterations in local enzyme requirements. As altered levels of catecholamine neurotransmitter have been implicated in the pathogenesis of a number of neurodegenerative and psychiatric disorders, such as Parkinson's disease, depression, anxiety disorders, and schizophrenia, it is important to understand how catecholamine neurotransmitter synthesis is regulated in the nerve terminal.
To identify factors that might regulate the presynaptic levels of catecholamines, we evaluated the hypothesis that TH mRNA is trafficked to the axons through localization sequences present in the 3′UTR and that it is locally translated. In this report, we provide the first direct evidence that TH mRNA is localized to axons of sympathetic neurons, and demonstrate that its local translation facilitates axonal dopamine synthesis. Moreover, we show that the 3′UTR of TH mRNA contains information for its axonal trafficking. Taken together, these results suggest that catecholamine synthesis is locally regulated in the axons and presynaptic nerve terminal.
RESULTS
Tyrosine hydroxylase mRNA is present in the distal axons of sympathetic neurons
To test the hypothesis that TH is locally synthesized in axons, we first examined whether the mRNA encoding this enzyme is present in distal axons. Toward this end, we cultured dissociated SCG neurons in compartmentalized Campenot chambers (Fig. 1A). After 7 DIV, total RNA was isolated from distal superior cervical ganglion (SCG) axons growing in the side compartments of Campenot chambers and, as a positive control, from the neuronal cell bodies plated in the central compartment of the chambers. Although we routinely monitor the side compartments of the Campenot cell culture chambers, where the distal axons of SCG neurons are localized, for the presence of neuronal cell soma and nonneuronal cells using light microscopy, trace cellular contamination of the axonal RNA preparation might still be detectable at the level of resolution afforded by microarray analysis or real-time PCR methodology. To address this issue, the purity of the RNA samples prepared from the distal axons was assessed by RT-PCR using gene-specific primer sets for mRNAs present in neuronal cell soma. As shown in Figure 1B, amplicons for Cholinergic receptor, nicotinic, α5 (Chrna5), and Ras-related GTP binding B (RragB), were readily visualized in RNA prepared from the neurons present in the central compartment, but were not observed in RNA obtained from distal axons. This finding was consistent with previous results, which established the purity of the axonal mRNA preparations (Natera-Naranjo et al. 2010).
FIGURE 1.
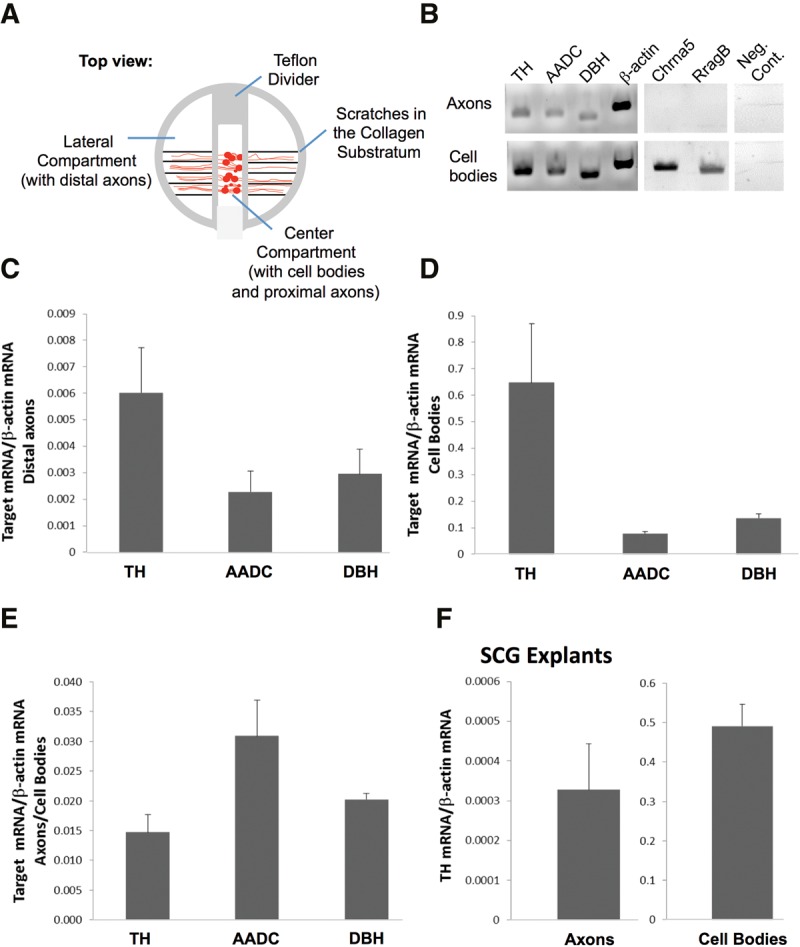
mRNA encoding TH, AADC, and DBH localize to distal SCG axons. (A) Schematic diagram showing the multicompartment Campenot cell culture chamber (top view). Dissociated rat superior cervical ganglion (SCG) neurons are plated in the center compartment and contain the cell bodies and proximal axons. Axons grow into the lateral compartments, devoid of neuronal or glial cells. (B–E) qRT-PCR analyses performed on total RNA obtained from distal axons of dissociated primary SCG neurons grown in Campenot chambers (7 DIV). RT-PCR-based (B) and qPCR-based mRNA quantification of catecholaminergic enzymes relative to β-actin (C–E) are shown. TH, tyrosine hydroxylase; AADC, aromatic l-amino acid decarboxylase; DBH, dopamine-β hydroxylase; cholinergic receptor, nicotinic, α5; RragB, Ras-related GTP binding B. (F) qRT-PCR analysis performed on total RNA obtained from distal axons of SCG explants from 3-d-old rat pups cultured in Campenot chambers. Error bars are SEM from at least three independent experiments.
The presence of axonal TH mRNA could be readily detected by RT-PCR from RNA isolated from distal axons devoid of any cell body mRNAs (Fig. 1B). Similarly, RT-PCR analysis showed that aromatic L-amino acid decarboxylase (AADC) and dopamine β-hydroxylase (DBH), the two enzymes that together with TH comprise the noradrenaline biosynthetic pathway, are also present in distal axons (Fig. 1B). In addition, mRNA encoding neuropeptide Y (NPY), a peptide coreleased with catecholamines from sympathetic nerve terminals, was also detected in axonal RNA using RT-PCR (data not shown). Using quantitative RT-PCR, we examined the relative quantity of TH, AADC, and DBH mRNA in the axons and cell bodies of SCG neurons. This study showed that TH mRNA levels were most abundant among these enzymes (relative quantity axons: TH: 0.0060 ± 0.0017, AADC: 0.0023 ± 0.0008, DBH: 0.0030 ± 0.0009; relative quantity cell body: TH: 0.6481 ± 0.2221, AADC: 0.0779 ± 0.0071, DBH: 0.1357 ± 0.0164). To eliminate possible in vitro artifacts associated with the culture of dissociated primary neurons, we performed qRT-PCR analyses using total RNA extracted from axons of whole-SCG tissue explants grown in Campenot chambers. The results of these experiments established that TH mRNA was also present in axons elaborated from SCG tissue explants (Fig. 1F).
To confirm that TH mRNA is present in axons, we used in situ hybridization histochemistry. In these experiments, the hybridization signal appeared as discrete puncta along the entire length of the axon bundles (Fig. 2A, upper panel). In contrast, no signal was detected in axon bundles incubated with a riboprobe against the bacterial mRNA DapB, used as a negative control (Fig. 2A, middle panel). In addition, the hybridization signal for the mRNA encoding an endoplasmic reticulum localized protein, Peptidylprolyl isomerase B (Ppib), was restricted to the cell body, indicating that the TH signal detected in axons was specifically due to local transport of TH mRNA (Fig. 2B, lower panel). Similar results were obtained with dissociated SCG neurons cultured on glass chamber slides. Again, the hybridization signal for TH appeared as discrete puncta along the entire length of single axons, while no signal was detected in axons using the negative control probe (Fig. 2B). Interestingly, a subset of TH mRNA fluorescent puncta appeared to localize in regions of axonal varicosities, suggesting that TH mRNA may accumulate in synaptic boutons. To establish the nature of these varicosities, we performed immunostaining of SCG axon bundles with TH protein and synapsin (Fig. 2C) and with a synaptic vesicle-specific fluorescent dye, FM1–43 (data not shown). The results of these experiments confirmed that the observed axonal varicosities are synaptic boutons and showed that a large fraction of TH protein colocalizes in these regions (Fig. 2C). To test the hypothesis that a fraction of the neuronal TH mRNA population is present juxtaposed to axonal varicosities, we performed in situ hybridization for TH mRNA followed by immunostaining with synaptotagmin protein. As shown in Figure 2D, TH mRNA hybridization signals colocalize with synaptotagmin, suggesting that TH mRNA accumulates in or is adjacent to synaptic boutons.
FIGURE 2.
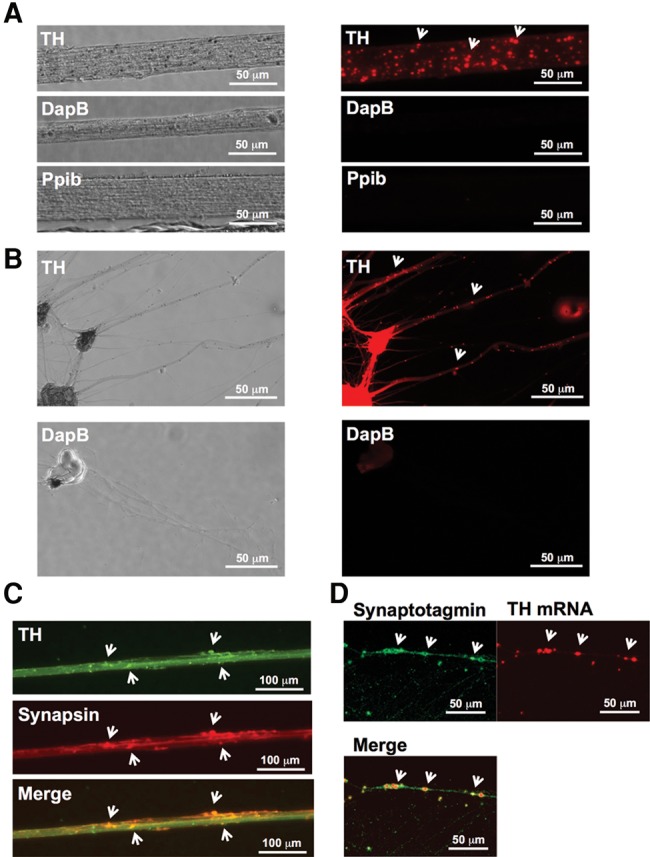
TH mRNA can be visualized in the distal axons of SCG neurons. (A) Fluorescent in situ hybridization analysis of axon bundles from primary SCG neurons grown in Campenot chambers. TH mRNA appears as discrete puncta along axon bundles (arrows). The mRNA for the bacterial gene DapB (negative control) is not detectable. mRNA for Ppib, an endoplasmic reticulum-associated protein, is not localized to the axons. (B) RNAscope analysis of single axons from dissociated primary SCG neurons grown in monolayer cell culture. In axons hybridized with a TH-specific riboprobe, TH mRNA appears as discrete puncta (arrows). In axons hybridized with the control riboprobe DapB, no signal is detectable. (C) Visualization of TH and synapsin protein in axon bundles from primary SCG neurons. (D) Visualization of synaptotagmin protein and TH mRNA in axons of dissociated SCG neurons. Note that synaptotagmin appears to colocalize with TH mRNA.
TH is synthesized in distal axons of primary noradrenergic SCG neurons
The finding that TH mRNA is localized to distal axons suggests that the transcript may be locally translated. To evaluate this hypothesis, we tested first whether TH mRNA associates with the translational apparatus of the axon. Polysomes were isolated from the distal axons of SCG neurons and the presence of TH-encoding mRNA determined by qRT-PCR methodology using gene-specific primers for TH. Results of this experiment revealed the presence of TH mRNA in RNA isolated from axonal polysomes (Fig. 3). Addition of ethylenediaminetetraacetic acid (EDTA), a chelating agent known to dissociate polysomes, markedly diminished the PCR-generated signals. Under these experimental conditions, the dissociated polysomes are incapable of sedimenting through 2.0 M sucrose and hence were eliminated from the polysome fraction (Giuditta et al. 1991; Crispino et al. 1993, 1997). Less than 2% of total TH RNA co-sedimented with the polysome fraction in the presence of EDTA.
FIGURE 3.
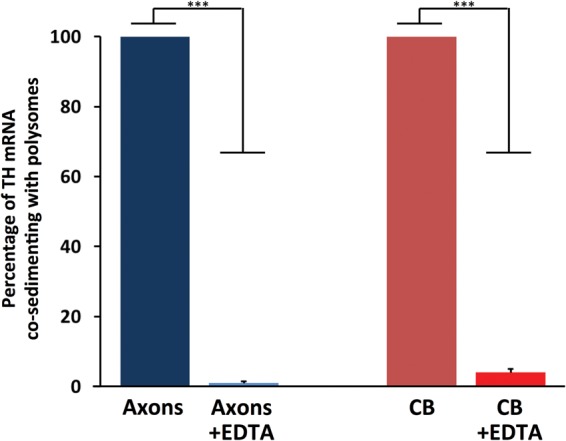
TH mRNA is present in axonal polysomes. qRT-PCR analysis of polysomal RNA from axons or cell body of SCG cell cultures (7 DIV). The histogram shows the percentage of TH mRNA that co-sediments with polysomes in polysomes isolated in the presence of EDTA relative to polysomes isolated in the absence of EDTA Treatment of polysomes with EDTA significantly decreased the levels of TH RNA bound to polysomes, demonstrating the purity of the isolated polysome fraction. CB, polysome-bound TH RNA from cell bodies; CB + EDTA, polysome-bound TH RNA from cell bodies + EDTA treatment. Data are mean ± SEM from three independent experiments. Student's t-test, (***) P ≤ 0.001.
To further evaluate the hypothesis that TH mRNA is translated locally in the axon, we used bio-orthogonal noncanonical amino acid tagging (BONCAT) to metabolically label newly synthesized proteins with the methionine analog, L-azidohomoalanine (AHA) (Dieterich et al. 2006). In this experimental approach, proteins synthesized during the labeling period incorporate AHA instead of methionine, and labeled protein can be biotinylated and subsequently isolated from the proteome by affinity purification. Axons growing in the side compartments of Campenot chambers were incubated in methionine-free medium containing AHA for 6 h, and AHA-labeled proteins were cross-linked to biotin and affinity purified with streptavidin beads. Western analysis of AHA-labeled proteins clearly showed that TH is locally synthesized in the axon (Fig. 4A). In contrast to the results obtained from axonal lysates, no signal above background was detected in affinity-purified proteins isolated from the parental cell soma of axons labeled with AHA for 6 h (Fig. 4A). This result supported the finding that the AHA-labeled TH detected in axons was locally synthesized. To further exclude the possibility that AHA-labeled TH was transported to the axon from the parental cell soma during the 6-h labeling period, newly synthesized proteins in the cell body were labeled with AHA for 6 h. Newly synthesized, biotinylated TH was then identified by Western blotting of affinity-purified protein obtained from both the cell body and distal axons. Although AHA-labeled TH was detected in the protein isolated from the cell bodies (i.e., positive controls), no AHA-labeled TH was observed in the distal axons (Fig. 4B). This finding confirmed that the AHA-labeled TH in the axon does not accrue from the cell body at least during the 6-h metabolic labeling period used in these experiments. In a third experiment, axons were severed from their cell soma and subsequently exposed to culture media containing AHA for 6 h. In this experiment, AHA-labeled TH was readily detected in the protein lysates isolated from the severed axons, confirming that TH can be locally synthesized from endogenous TH mRNA (Fig. 4C).
FIGURE 4.
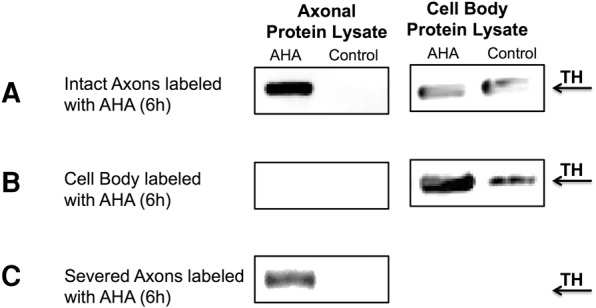
TH is locally synthesized in distal axons of SCG neurons. (A) Western analysis showing newly synthesized TH protein in SCG axons labeled with AHA for 6 h. (B) Western analysis of AHA-labeled proteins from cell bodies of 7 DIV SCG cultures labeled with AHA for 6 h. Note that newly synthesized TH is not transported from the cell body to the axon within the 6-h labeling period. (C) Western analysis showing newly synthesized TH protein in SCG axons labeled with AHA for 6 h in axons severed from the cell body prior to AHA labeling. AHA, affinity-purified proteins from axons or cell body incubated with AHA for 6 h. Control, affinity-purified proteins from axons or cell bodies not incubated with AHA.
Local synthesis of TH facilitates axonal dopamine production
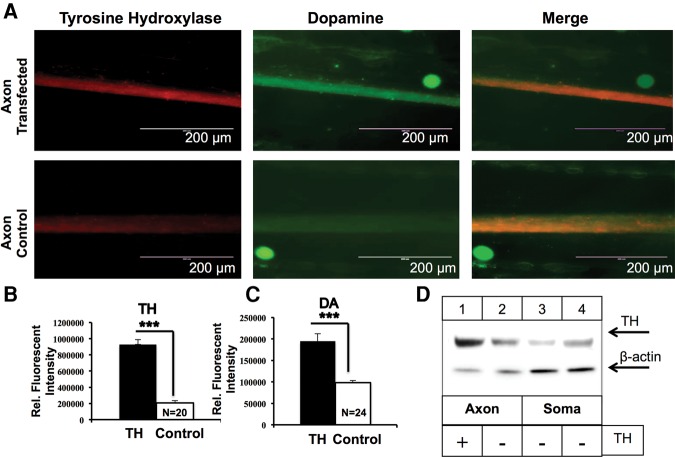
Since TH is the rate-limiting enzyme in catecholamine biosynthesis, we examined whether local synthesis of TH affects dopamine synthesis in the axons of SCG neurons. Toward this end, TH mRNA was transfected into the lateral compartments of Campenot chambers, harboring the distal axons of SCG neurons and TH and dopamine levels were assessed using immunocytochemistry. Transfection with TH mRNA resulted in a 4.5-fold increase in axonal TH protein levels compared with the endogenous TH levels in sham-transfected axons (relative fluorescent level TH in transfected axons: 97764.85, relative fluorescent level TH in sham-transfected axons: 21877.76, P < 0.0001) (Fig. 5A,B). To explore whether TH overexpression results in enhanced dopamine levels in the distal axons of SCG neurons, we monitored dopamine levels 24 h after transfecting the distal axons with TH mRNA. Dopamine levels increase by ≈50% in distal axonal compartments when compared with dopamine levels in sham-transfected axons (relative fluorescent level dopamine in transfected axons: 19356.7, relative fluorescent level dopamine in sham-transfected axons: 9768.6, P < 0.0001; Fig. 5A,C). Western blot analysis of TH protein in axonal and parental cell soma 24 h after transfection of distal axons with TH mRNA showed that TH overexpression was restricted to distal axons, with no TH overexpression observed in parental cell soma of axons transfected with TH (Fig. 5D).
FIGURE 5.
Local translation of TH mRNA facilitates axonal dopamine synthesis. (A) Intra-axonal TH and dopamine levels were measured using immunocytochemistry in axons lipofected with TH mRNA (upper panel) or sham-transfected (lower panel). Increased dopamine levels are detected in axons locally overexpressing TH. (B,C) Fluorescence intensity as a measure of TH (B) and dopamine (C) levels was quantified using ImageJ, and fluorescence levels are indicated as relative fluorescence intensity. Data are mean ± SEM from the measurement of 20–24 axons from three independent experiments. Student's t-test, (***) P ≤ 0.0001. (D) Western-blot analysis of axonal and cell body protein lysates from SCG axons transfected with TH mRNA or transfection reagent. β-actin was used as a loading control. Blots of axonal and cell body TH proteins showed significant overexpression of TH protein levels in axons transfected with TH mRNA, whereas TH levels in parental cell soma from transfected axons remained at basal levels.
The 3′UTR of TH mRNA mediates reporter mRNA trafficking to axons
mRNA trafficking and localization have been shown to be directed by cis-acting elements often found in the 3′UTR of the mRNA (Martin and Zukin 2006; Jambhekar and DeRisi 2007; Aschrafi et al. 2010). Comparative sequence analysis of the 3′UTR of TH revealed that there are several evolutionary conserved regions that may contain regulatory elements (Fig. 6A). RNA secondary structure analysis using Mfold (Zuker 2003) and RNA-fold (Gruber et al. 2008) indicated that the TH 3′UTR encompasses three putative stem–loop structures (Fig. 6B). Among these structures, stem–loop III (nucleotides 213–238) manifested ∼73% sequence homology across three mammalian species. Interestingly, cis-acting localizing elements have been shown to often form stem–loop structures recognized by RNA binding proteins (Bassell and Singer 1997; Kiebler and Bassell 2006). Based on these observations, it was hypothesized that the 3′UTR of TH contained elements that mediated the localization of TH mRNA to distal axons and presynaptic terminals. To test this postulate, we transfected primary SCG neurons, cultured in Campenot chambers with a plasmid containing the CMV promoter, the open reading frame (ORF) of tandem dimer (td) Tomato red fluorescent protein, and the entire 3′UTR of TH (Full 3′UTR, Fig. 6C). In the negative control, neurons were transfected with a tdTomato plasmid containing the SV40 3′UTR (Fig. 6C). Importantly, reporter transcripts containing the SV40 3′UTR have been shown to be restricted to the cell bodies of primary neurons, and have previously been used as negative controls in studies of axonal localization of mRNAs (Goetze et al. 2003; Aschrafi et al. 2012).
FIGURE 6.
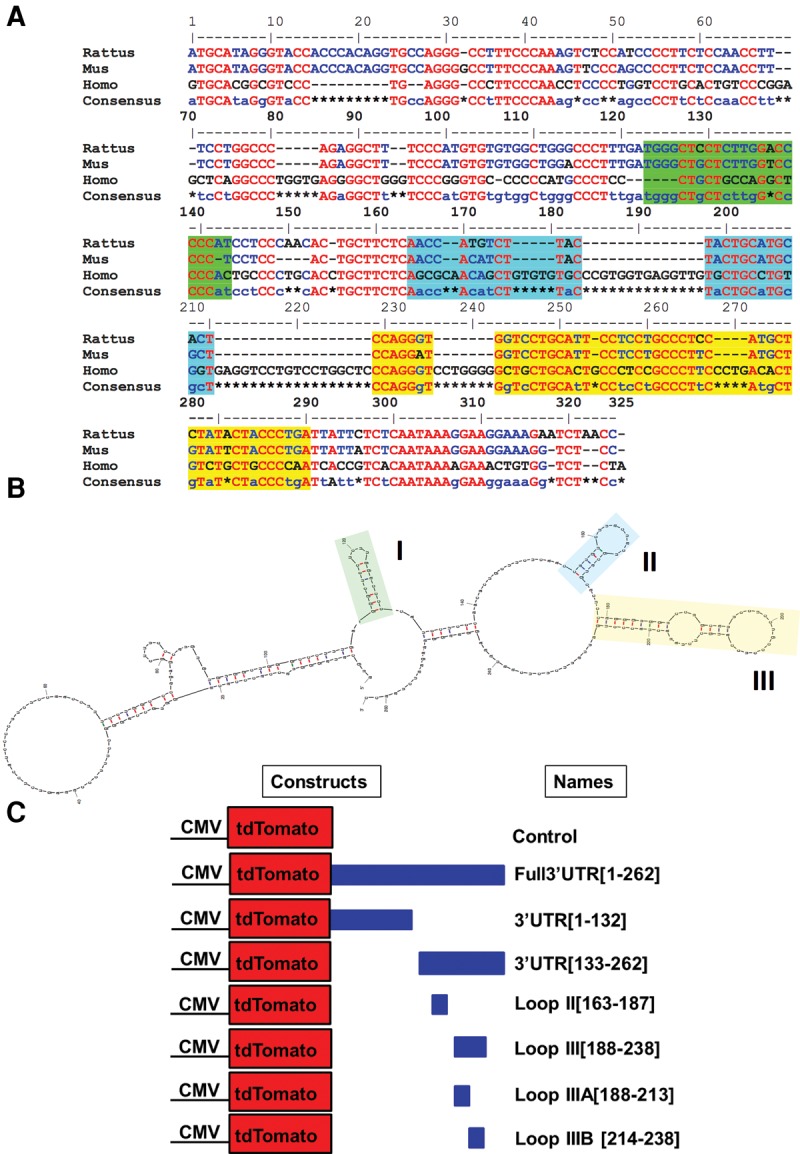
The 3′UTR of mammalian TH mRNA is highly conserved and contains three putative stem–loop structures. (A) Sequence alignment of the 3′UTR of TH mRNAs from three mammalian species shows a high degree of sequence conservation. The red and blue letters represent nucleotides conserved among all three and in two out of the three species, respectively. Green, blue, or yellow highlighted regions correspond to the sequences of the three putative stem–loop structures. (B) Structure of the TH 3′UTR, as determined by secondary structure prediction analysis (Mfold) (Zuker 2003). Yellow denotes a hairpin stem–loop (50 nt), which is highly conserved. (C) Schematic of plasmid constructs used to transfect SCG neurons. The control plasmid was used unmodified as acquired from Clontech and contains a SV403′UTR. In the Full 3′UTR [1–262 bp] plasmid construct, the full-length 3′UTR of TH was inserted following the tdTomato ORF.
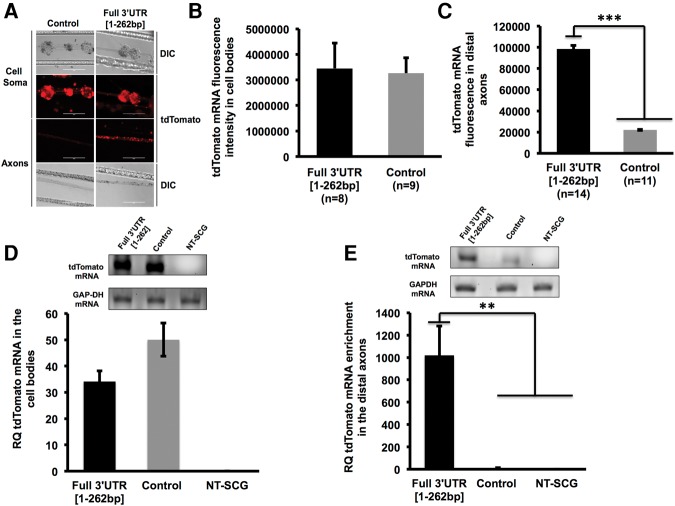
The vectors expressing the chimeric reporter mRNA were introduced into SCG neurons (6 DIV) by transfecting the neuronal cell bodies and proximal axons located in the center compartment of Campenot chambers. On DIV 9, the cultures were fixed and processed for RNA in situ hybridization using a tdTomato-specific riboprobe. In neurons transfected with the Full 3′UTR [1–262 bp] construct, the hybridization signal appeared as discrete puncta situated along the entire length of the distal axon (Fig. 7A). This pattern of reporter mRNA signal was similar to the pattern observed for endogenous TH mRNA (see Fig. 2A,B). In contrast, the hybridization signal derived from the control plasmid was almost exclusively restricted to the cell soma. Measurement of fluorescence pixel intensity in the distal axons showed nearly a fivefold increase in the intensity of tdTomato mRNA signal in axons of neurons transfected with the plasmid containing the Full TH 3′UTR compared to the control plasmid (Fig. 7C). qRT-PCR analysis of RNA isolated from distal axons confirmed that higher levels of the reporter mRNA were present in distal axons (Fig. 7E) when neurons were transfected with the Full 3′UTR [1–262 bp] plasmid rather than the control plasmid (Fig. 7D). Worthy of note, both in situ and qRT-PCR analysis showed that the levels of the reporter mRNA in the cell soma did not differ significantly between experimental and control reporter constructs (Fig. 7B,D). These results indicate that the higher levels of tdTomato mRNA in the axons of neurons transfected with Full 3′UTR [1–262 bp] are not due to differences in transfection efficiency or expression levels between the experimental and control plasmids. In addition, these results demonstrate that the TH 3′UTR contains a cis-acting regulatory element that is both necessary and sufficient for the targeting of TH mRNAs to the distal axons.
FIGURE 7.
The 3′UTR of TH mRNA is sufficient to target reporter mRNA into distal axons. (A) Representative DIC and fluorescent images of reporter (tdTomato) mRNA in the cell bodies and distal axons of SCG neurons as visualized by in situ hybridization. tdTomato-specific fluorescent in situ hybridization signal appears as punctate mRNA granules in the distal axons. A significant amount of staining was detected in the axons of cell bodies transfected with the tdTomato construct containing the TH Full 3′UTR as compared to the control. The axons labeled with a negative control probe DapB (i.e., Bacillus subtilis dihydrodipicolinate reductase gene) produced little or no fluorescence under identical hybridization and wash conditions (data not shown). This probe was used as a negative control because it is a bacterial transcript not present in rat tissue. Scale bar, 200 µm. (B,C) Quantification of reporter mRNA fluorescence in situ hybridization signal in cell bodies (B) and distal axons (C) of SCG transfected neurons. Note that the signal intensity in distal axons of neurons transfected with Full 3′UTR [1–262 bp] is about five times that of distal axons of neurons transfected with the control plasmid, whereas the signal in transfected cell bodies do not differ. (D) tdTomato mRNA levels in the cell bodies of SCG neurons determined by qRT-PCR 72 h after DNA transfection. GAPDH served as an internal control. (E) tdTomato mRNA enrichment levels in the distal axons of SCG neurons 72 h after plasmid DNA transfection. GAPDH served as an internal control. All experiments were repeated three times with similar results. Histograms shown are representative of one of these experiments. Cell soma and axon tdTomato mRNA levels were visualized by fluorescence microscopy and quantified using Image J. Data are normalized to the fluorescence observed in nontransfected neurons. Each experiment was repeated three or more times with similar results. Error bars represent the standard error of the mean. Two-tailed Student's t-test (***) P ≤ 0.0001, (**) P ≤ 0.001. NT-SCG, nontransfected SCGs.
The 3′UTR of TH mRNA contains an axonal targeting element (25 nt)
To identify sequences in the 3′UTR of TH mRNA required for axonal targeting of the transcript, we conducted a series of deletion experiments by transfecting SCG neurons with constructs encoding the tdTomato ORF followed by selected segments of the 3′UTR of TH mRNA.
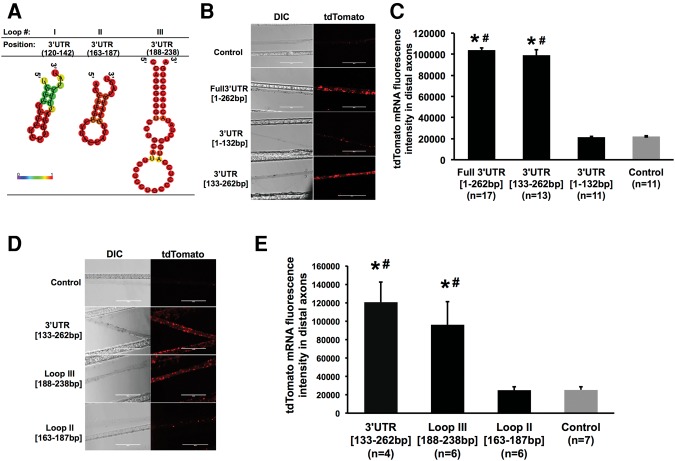
First, we observed that two of the three stem–loop structures (predicted by secondary structure analyses) to be present in the 3′UTR are located in the final 130 nt of the region (Fig. 8A). Based upon this observation, we generated chimeric plasmids encoding the tdTomato ORF followed by either the first 132 nt of the TH 3′UTR, 3′UTR [1–132 bp] or the final putative stem–loop rich 130-nt 3′end, 3′UTR [133–262 bp]. In neurons transfected with the constructs containing either the full-length TH 3′UTR or the 3′UTR [133–262 bp] reporter, RNA hybridization signal was clearly detectable in distal axons (Fig. 8B). In addition, the axonal distribution of 3′UTR [133–262 bp] was not significantly different from that of Full 3′UTR [1–262 bp] (Fig. 8C). In contrast, in neurons transfected with the construct containing the first 132 nt of the TH mRNA 3′UTR or the control vector, the reporter RNA hybridization signal was undetectable in distal axons (Fig. 8B,C). These experiments raised the possibility that the 3′ end (nucleotides 133–262) of the TH 3′UTR containing two putative hairpin stem–loops (stem–loops II and III) may mediate the localization of the tdTomato reporter mRNA to the distal axons of SCG neurons.
FIGURE 8.
A putative 50-nt stem–loop in 3′UTR of TH mRNA is sufficient to target a reporter gene to the axon. (A) Representative graphic depictions of potential stem–loop structures in the 3′UTR of TH predicted by RNA-fold; color scale denotes base pair probabilities. (B) Representative DIC and fluorescent images of reporter mRNA as visualized by in situ hybridization in distal axon bundles of SCG neurons transfected with Full3′UTR [1–262 bp], 3′UTR [1–132 bp], 3′UTR [133–262 bp], or the control plasmid. Scale bar, 200 µm. (C) Quantification of fluorescence in situ hybridization signal of tdTomato mRNA in axons. (*) P < 0.0001 versus control; (#) P < 0.0001 versus 3′UTR [1–132 bp]. (D) In situ hybridization of SCG cultures transfected with tdTomato vectors containing the final 130 nt, the Loop II [163–187 bp], Loop III [188–238 bp] sequences, or the control plasmid. Scale bar, 200 µm. (E) Quantification of fluorescence in situ hybridization signal intensity for tdTomato mRNA in axon bundles. One-way ANOVA with Bonferroni corrected post-hoc t-test to compare between the experimental groups. (*) P < 0.01 versus control; (#) P < 0.01 versus Loop II [163–187 bp]. All experiments were repeated three times with similar results. Histograms shown are representative of one of these experiments. Error bars represent the standard error of the mean.
To further delineate the identity of the localization element, we created chimeric plasmids encoding tdTomato ORF followed by the sequence of either of the two stem–loops predicted to be present in the last 130 nt of the TH 3′UTR. Neurons were transfected with either of the following plasmids: 3′UTR [133–262 bp], Loop II [163–187 bp], Loop III [188–238 bp], or control. As shown in Figure 8D,E, transfection with Loop III [188–238 bp] resulted in significantly higher amounts of tdTomato mRNA in distal axons compared to stem–loop II [163–187 bp] or the control plasmid. These experiments revealed that the stem–loop structure III contained elements requisite for the trafficking of tdTomato chimera mRNAs to distal axons.
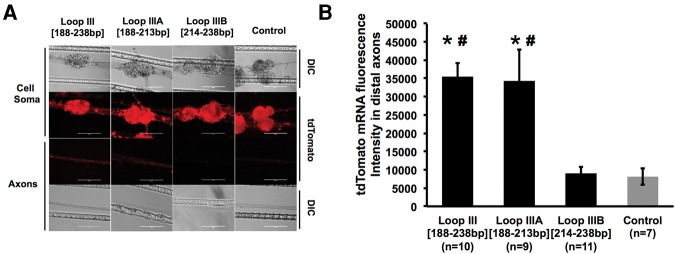
Finally, to assess whether the identified Loop III represents the minimal functional element for axonal trafficking of the message, tdTomato vectors were created encoding the tdTomato ORF followed by the initial 25 nt of stem–loop III (Loop IIIA [188–213 bp]) or the final 25 nt of loop III (Loop IIIB [214–238 bp]). Subsequently, SCG somata were transfected with these vectors to determine whether these fragments had the capacity to transport tdTomato mRNA to distal axons. As shown in Figure 9A, minimal reporter fluorescence was observed in axons that were transfected with plasmid DNA that contained the Loop IIIB [214–238 bp] sequence. However, axons transfected with Loop IIIA [188–213 bp] constructs showed similar fluorescence signal as compared to the intact Loop III [188–238 bp] constructs. Together, these data suggest that TH mRNA localization depends on the recognition of the Loop IIIA [188–213 bp] 25-nt sequence element (Fig. 9B).
FIGURE 9.
A 25-nt putative stem–loop structure is the minimal element requisite for reporter mRNA localization. (A) Representative DIC and fluorescent images of reporter mRNA as visualized by in situ hybridization in distal axon bundles of SCG neurons transfected with Loop III [188–238 bp], Loop IIIA [188–213 bp], or Loop IIIB [214–238 bp]. Scale bar, 200 µm. (B) Quantification of fluorescence in situ hybridization signal intensity for tdTomato mRNA in axon bundles of neurons transfected with Loop III [188–238 bp], Loop IIIA [188–213 bp], or Loop IIIB [214–238 bp]. One-way ANOVA with Bonferroni corrected post-hoc t-test to compare between the experimental groups. Error bars represent the standard error of the mean. (*) P < 0.001 versus control; (#) P < 0.01 versus Loop IIIB [214–238 bp]. The experiment was repeated three times with similar results. Histogram shown is representative of one of these three experiments.
DISCUSSION
In this report, we provide, for the first time, direct evidence for the axonal trafficking and local translation of TH mRNA in axons of sympathetic neurons. These findings derive from several independent experimental approaches.
First, we show that TH mRNA is localized to distal axons. By growing dissociated SCG neurons in compartmentalized culture chambers we were able to extract pure axonal RNA (Natera-Naranjo et al. 2012). This RNA sample was used to drive qRT-PCR analyses that showed that TH mRNA is present in the RNA obtained from distal axons. The results of in situ hybridization experiments showed that numerous punctate hybridization signals were distributed along the entire length of the axon. Interestingly, the punctate hybridization signals seemed to colocalize in the axon juxtaposed to synaptic boutons (i.e., axo-axonal synapses). To establish the identity of these varicosities, we stained axons with presynaptic nerve terminal markers, such as synapsin and synaptotagmin. Costaining for TH protein also showed that TH accumulates in these structures. Interestingly, the results of colocalization experiments using in situ hybridization followed by immunocytochemistry suggest that TH mRNA also accumulates in association with these structures (i.e., synaptic boutons).
Second, we demonstrate that TH mRNA is present in axonal polysomes. In this regard, we lysed distal axons growing in the side compartments of Campenot chambers and purified axonal polysomes by sedimentation through 2 M sucrose. This technique has been used previously to isolate translationally active axonal polysomes (Giuditta et al. 1991; Crispino et al. 1997). The finding derived from qRT-PCR analysis clearly established that TH mRNA is present in axonal polysomes. To ensure that the amplicons generated by qRT-PCR were indeed derived from polysomal RNA, we purified polysomes with an isolation buffer containing EDTA, a Mg2+ chelating agent known to dissociate polysomes. Under these isolation conditions, the dissociated polysomes are unable to sediment through heavy sucrose (Giuditta et al. 1991; Crispino et al. 1993). The qRT-PCR signal generated by amplification of TH greatly diminished in the presence of EDTA, providing additional evidence that TH mRNA associated with polysomes.
Third, we show that TH mRNA is locally translated in SCG axons. Although the in vitro translation experiment showed that TH mRNA associates with polysomes in vitro, it did not demonstrate that translation occurs in the axons under the culture conditions used in these experiments. To address this issue, we labeled axonally synthesized proteins with AHA, a methionine analog. Incorporation of this modified amino acid into newly synthesized protein allowed for its subsequent biotinylation and immunopurification of newly synthesized proteins from the cell lysate using streptavidin-coated magnetic beads. In these studies, TH was clearly detectable by Western analysis conducted on AHA-labeled proteins extracted from the distal axons. However, in these initial experiments, we could not rule out the possibility that the AHA-labeled TH detected in axons had actually been synthesized in the cell bodies and diffused or had been transported from the parental cell body during the 6-h labeling period. To exclude this possibility, we conducted three sets of experiments. First, we lysed the parental cell bodies of axons exposed to AHA and conducted affinity purification and Western analysis on the cell body lysate: Little immunoreactive signal was detected above background. Second, we labeled the cell body directly with AHA and conducted Western analysis on the lysates obtained from the distal axons. Again, no detectable signals were obtained from distal axons, while we were able to detect newly synthesized TH in the parental cell soma. Finally, we severed axons from their parental cell soma prior to incubation with AHA, so that there was no possibility of proteins being transported to axons. Indeed, AHA-labeled TH was detectable in the severed axons. Taken together, these results clearly demonstrate that TH is locally synthesized in SCG axons.
Last, in this report, we identify a sequence element in the 3′UTR of TH mRNA that directs TH mRNA localization to distal axons. In this regard, we provide data demonstrating that the axonal targeting of TH mRNA is mediated by regulatory elements (“zip-codes”) that are sufficient to traffic a reporter mRNA transcript to the axon. Bioinformatic analyses (Mfold/RNA-fold) of the rat TH 3′UTR predicted the existence of three putative stem–loop structural/functional motifs that may serve as cis-acting regulatory elements. Moreover, sequence alignment studies of mammalian TH homologs showed a high degree of conservation in residues that formed the putative stem–loop structures. These observations further suggested that the predicted hairpin loops may contain information for targeted delivery of the TH mRNA transcript.
Although it has been shown that structural motifs in the 3′UTR of RNA functions in promoting mRNA stability (Bevilacqua and Blose 2008), our study suggests that the fusion of the TH 3′UTR elevated reporter mRNA levels in the distal axons via alteration in mRNA transport rather than mRNA stability. These data are consistent with a critical role played by hairpin stem–loop secondary structures in mediating RNA trafficking (Chartrand et al. 1999; Jambhekar and DeRisi 2007; Aschrafi et al. 2010; Meer et al. 2012). Stepwise deletion analyses of the full TH 3′UTR suggests that an evolutionary conserved sequence located at the terminal end of the transcript functions as the minimal element (25 nt) for axonal transport in SCG neurons. When the full TH 3′UTR or 3′UTR [133–262 bp] segment of the 3′UTR were transfected into the cell somas, we observed elevated levels of tdTomato reporter in the distal axons of SCG neurons. Further deletion studies of 3′UTR [133–262 bp] suggested that formation in the 50-nt-long stem–loop sequence might be important for axonal mRNA trafficking. Disrupting the predicted 50-nt stem–loop by reducing the length of the sequence into two 25-nt fragments (Loop IIIA [188–213 bp] and Loop IIIB [214–238 bp]) resulted in marked reduction in reporter axonal mRNA transport in the neurons transfected with Loop IIIB [214–238 bp] construct. On the contrary, the expression levels of the reporter mRNA in axons of neurons transfected with Loop IIIA [188–213 bp] were not significantly different from those transfected with the intact 50-nt Loop III [188–238 bp] construct. This result suggests that Loop IIIA [188–238 bp] may represent the minimal element for TH mRNA transport/trafficking to the distal axons of SCG neurons. Interestingly, Mfold and RNA-fold secondary structure analyses of the Loop IIIA [188–213 bp] and Loop IIIB [214–238 bp] sequences showed that Loop IIIA [188–213 bp] also has the potential to form a stem–loop structure, while Loop IIIB [214–238 bp] does not. This finding suggests that secondary RNA structure may also contribute to the trafficking activity of the Loop IIIA fragment. However, further mutational and SHAPE analyses must be performed to confirm the validity of this hypothesis.
The expression pattern of the chimeric reporter mRNA was similar to the in situ hybridization pattern obtained for endogenous TH mRNA, which appears to accumulate juxtaposed to axonal varicosities. This observation raises the possibility that the 3′UTR of TH mRNA may not only direct axonal localization of the transcript, but might also contain a synaptic localization element. This finding is consistent with studies showing the presence of cis-acting localizing elements for specific neurite subcompartments in the untranslated regions of neuronal transcripts (Muslimov et al. 2004; Meer et al. 2012). Finally, in addition to directing axonal transport, the 3′UTR of TH mRNA may also regulate translation of the transcript via the binding of microRNAs (for review, see Kaplan et al. 2013). Further investigation will be needed to fully elucidate the role played by the 3′UTR of TH in the regulation of its axonal trafficking, local expression, and targeting of the mRNA to the presynaptic compartment of the neuron.
Our finding that TH mRNA is present in axons is consistent with the detection of TH mRNA in brain regions innervated by catecholaminergic terminals (for example, see Lewis et al. 1991; Melia et al. 1994). In the adult rat brain, the presence of TH mRNA transcripts was reported in the cerebellum, striatum, and pituitary neurointermediate lobe (NIL), all regions that are devoid of catecholamine-synthesizing cells, but receive significant catecholaminergic innervation (Melia et al. 1994). TH mRNA levels in these regions were increased after administration of reserpine, suggesting a functional role for the axonally localized TH transcript (i.e., restoration of depleted stores of catecholamine). This study, however, is the first to provide direct evidence for the presence and local translation of TH mRNA in axons and presynaptic nerve terminals.
The findings that TH is synthesized locally in axons and that the 3′UTR of TH contains an axonal localization signal suggest that the axonally localized TH mRNA plays an active role in neurotransmitter metabolism. However, the exact function of the TH transcript in axons remains to be elucidated. Given that TH protein has been shown to be anterogradely transported to the axon, it is reasonable to consider whether axons would need to locally synthesize TH. Although mRNA is generally present in axons at much lower levels than in the cell body, its translation may be required for axons to quickly and effectively respond to environmental cues. In addition, newly synthesized protein can have different physicochemical characteristics from “older” proteins. For example, Holt and colleagues have shown that local synthesis of β-actin is required for growth cone turning in response to netrin stimulation (Leung et al. 2006). Newly synthesized β-actin can polymerize more efficiently than “older” actin because of chaperone binding and protection from glutathionylation, a post-translational modification that decreases the rate of polymerization. Being the rate-limiting factor in the synthesis of catecholamines, TH expression and activity is highly regulated by both transcriptional and post-translational mechanisms (for review, see Lenartowski and Goc 2011). It would be interesting to compare the pattern of post-translational modification between newly synthesized and “older” TH. Future work could show whether the newly synthesized TH has enzymatic activity and whether axonally synthesized and cell body synthesized TH manifest differences in their pattern of post-translational modification. The fact that local overexpression of TH can augment dopamine levels in the axons suggests that newly synthesized TH is indeed enzymatically active.
Moreover, the mechanisms regulating the synthesis of dopamine at nerve terminals are only partially investigated (Galloway et al. 1986). Autoreceptor activation and end-product inhibition act as negative feed-back loop modulators of dopamine levels at nerve terminals of dopaminergic neurons. Previous studies reported on the presence of TH and other catecholaminergic enzymes in axon terminals of TH-positive neurons, suggesting that dopamine synthesis can take place at the nerve terminal (Kozicz 2001). Our study is the first to show that local expression of TH facilitates dopamine synthesis in distal axons. Dopamine production through localized TH synthesis at nerve terminals may provide an alternative mechanism for a rapid dopamine synthesis in response to nerve growth factors, or altered potassium levels. Future functional studies will delineate the physiological significance of axonal dopamine synthesis through locally translated TH.
In conclusion, our study provides the first direct evidence for the transport and local translation of TH mRNA in axons of SCG neurons and identifies a sequence element necessary and sufficient to traffic TH mRNA to the axon. Regulating the availability of the enzymes comprising the catecholamine biosynthetic pathway in the presynaptic nerve terminal might facilitate the activity-dependent modulation of local neurotransmitter synthesis and might ultimately lead to the development of new pharmacological strategies for the treatment of neuropsychiatric and for neurodegenerative disorders.
MATERIALS AND METHODS
Neuronal cell cultures
Superior cervical ganglia were removed from 3-d-old Harlan Sprague-Dawley rats and dissociated to single cell suspensions using gentleMACS Dissociator and Neuronal Tissue Dissociation Kit (Miltenyi Biotec). The dissociated cells were plated in the center compartment of three-chamber Campenot culture dishes (Hillefors et al. 2007) or, for purpose of imaging, on Nunc Lab-Tek II–CC2, glass chambers slides (Sigma-Aldrich). Cells were cultured in serum-free neurobasal medium (Life Technologies) supplemented with 2% B-27 supplement, 0.5 mM L-glutamine (Life Technologies), 25 µM glutamic acid (Sigma-Aldrich), 50 ng/mL NGF, 20 mM KCl, 20 U/mL penicillin, and 20 µg/mL streptomycin (Hyclone) for 3–14 d in vitro (DIV) prior to use. Medium was changed every 3–4 d. Two days after plating, 5-fluoro-2′-deoxyuridine (50 µM) was added to the culture medium to inhibit growth of nonneuronal cells. After 7–14 DIV, distal axons were harvested from the side compartments of culture chambers, which were devoid of neuronal soma or nonneuronal cells, as judged by phase-contrast microscopy.
RNA isolation and reverse transcriptase quantitative PCR (qRT-PCR)
RNA was isolated using Direct-Zol RNA MiniPrep (Zymo Reasearch) according to the manufacturer's instructions. Briefly, cells and proximal axons cultured in the central compartment of Campenot chambers or distal axons growing in the side compartments were lysed in 100 µL of TRIzol (Life Technologies). An equal volume of ethanol was added to the homogenate, and the mix was loaded onto a Zymo-Spin IIC column and washed with Direct-Zol RNA PreWash and Wash buffers. The RNA sample was eluted in 40 µL of water. On average, the concentration of the purified RNA was 2.5 ng/µL for RNA isolated from distal axons and 10 ng/µL for RNA isolated from neuronal cell bodies and proximal axons. Reverse transcription was performed mixing the purified RNA with qScript cDNA SuperMix (Quanta Biosciences) in a ratio of 5:1. The mix was incubated in a thermal cycler (MJ Research) for 5 min at 25°C, followed by 30 min at 42°C and 5 min at 85°C. For qRT-PCR analysis, 2 µL of cDNA (4 ng distal axon RNA, 16 ng for cell body and proximal axon RNA) was added to 10 µL SYBR green and 2 µL of QuantiTect primers (Qiagen). For each experimental sample, the reaction was run in triplicate, using a StepOne Real-Time PCR system (Applied Biosystems). The results were analyzed using the StepOne Software (Applied Biosystems), normalizing target mRNA levels to ribosomal protein subunit 18 (RPS18) or β-actin mRNA levels. All gene primers used were QuantiTect validated primers, except for the primers used for Chrna5 and RragB: Rat Chrna5, forward 5′-AACATCCACCACCGCTCTTC, rat Chrna5, reverse 5′-ATCTTCAACAACCTCGCGGAC, rat RragB forward 5′-AAAGAACAGCGAGATGCCCA, rat RragB reverse 5′-GCTGCAGAAGGAATGGATGG.
In situ hybridization histochemistry
In situ hybridization was performed on freshly fixed tissue using the RNAscope 2.0 High Definition (HD)–RED Assay (Advanced Cell Diagnostics) according to manufacturer's instructions. Cultured neurons were fixed in 10% formalin for 30 min, dehydrated in increasing concentrations of ethanol (50%, 70%, and 100%) and rehydrated (70% ethanol, 50% ethanol, PBS) before applying Pretreat 1 and Pretreat 3 reagents. A target specific riboprobe (rat TH, RefSeq: NM_012740.3; TdTomato, GenBank: EU855182.1) or control riboprobes (DapB of Bacillus subtilis strain, GenBank: EF191515.1; rat Ppib, RefSeq: NM_022536.2) were then hybridized for 2 h at 40°C, followed by incubation with signal amplification reagents. The hybridized riboprobe was visualized with RED2 under either bright field or fluorescence microscopy using a Texas Red filter. Worthy of note, tdTomato protein fluorescence does not persist after processing of samples for in situ hybridization so the hybridized riboprobe could be visualized using fluorescence microscopy under a Texas Red filter.
Immunocytochemistry
Neurons were fixed using 4% PFA for 30 min. After washing, permeabilization was accomplished with 0.3% Triton X-100 in PBS for 3 min followed by blocking in 3% bovine serum albumin (BSA) in PBS for 30 min at room temperature (RT) and incubation in primary antibody diluted in 3% BSA-PBS for 4 h at RT or overnight at 4°C. After incubation with the primary antibodies (anti-TH antibody CST 2792, 1:1000; anti-dopamine antibody Abcam [ab 1001], 1:1000), neurons were washed in PBS, incubated in secondary antibody for 1 h at RT and visualized using confocal microscopy. For combined in situ hybridization–immunocytochemical analysis, neurons were fixed and processed as described in the paragraph above. After the signal detection step with RED2, neurons were incubated with Triton-X 100 and processed for immunocytochemistry.
Metabolic labeling of newly synthesized proteins by bio-orthogonal noncanonical amino acid tagging (BONCAT)
Metabolic labeling of newly synthesized protein was performed using Click-iT L-azidohomoalanine (AHA) for Nascent Protein Synthesis Kit and Click-iT Biotin Protein Analysis Detection Kit (Invitrogen) according to manufacturer's instructions. Either axons located in the side compartments of Campenot chambers or neuronal somas located in the central compartment were incubated for 6 h with AHA in methionine-free culture medium and lysed in 50 mM Tris–HCl, pH 8.0, 1% SDS. AHA-incorporated protein was conjugated to biotin and the newly synthesized biotinylated protein was subsequently absorbed to streptavidin-coated magnetic beads (Pierce). The beads were washed five times with PBST (PBS + 0.1% Tween 20) and protein was eluted in 0.1% SDS by boiling for 5 min. Newly synthesized protein was quantitated by Western analysis, using polyclonal antibody specific for rat TH. Protein lysates from axons or cell bodies not incubated with AHA were used as controls. For each experiment, protein obtained from three culture dishes was combined prior to analysis.
Western analysis
Axon or soma protein lysates were boiled in LDS sample buffer and separated on NuPAGE Novex 4–12% Bis–Tris Gels (Life Technologies). Proteins were transferred to Hybond-LFP PVDF membranes (GE Healthcare) and membranes were blocked with ECL Advance Blocking Agent (GE Healthcare) for 1 h, followed by overnight incubation at 4°C with antibodies against TH (Cell Signaling, dilution 1:1000) or β-actin (Cell Signaling, dilution 1:1000). Membranes were washed in 1× TBS-T (1× TBS, 0.1% Tween 20) and incubated with horseradish peroxidase-labeled secondary antibody for 1 h at room temperature. After washing, membranes were developed with ECL Advance Western Blotting Detection Kit reagents, and imaged using Kodak image station 440. Band intensity was quantified using NIH image J Software.
Polysome preparation
Distal axons and their parental cell bodies were removed from the culture plates in an isolation buffer containing: 0.32 M sucrose, 50 mM Tris–HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 10 U/mL RNasin (Promega), 25 µg/mL tRNA, and 1 µg/mL cycloheximide, and tissue disrupted by homogenization in a glass/Teflon hand-held homogenizer. The isolation buffer was subsequently made 0.5% (v/v) with respect to both sodium deoxycholate and Triton X100, and clarified by centrifugation (13,000g for 10 min). Polysomes were collected by sedimentation through a 2.0 M sucrose cushion (100 µL) at 40,000 rpm for 4 h at 4°C (Beckman SW 50.1 rotor). In some experiments, polysomes were disaggregated prior to sedimentation by replacing the Mg2+ in the isolation buffers with the chelating agent, EDTA (20 mM). Under these conditions, polysomes are known to disassemble into the component ribosomal subunits, releasing their mRNA and nascent polypeptides (for example, see Giuditta et al. 1991). EDTA-treated polysomes are unable to sediment through the dense sucrose cushion. To obtain total polysomal RNA, polysome pellets were resuspended in TRIzol (Life Technologies), and RNA isolated using the Direct-zol RNA Mini Prep Kit (Zymo Research Corp) according to the manufacturer's instructions.
Constructs and SCG neuron transfection
All tdTomato chimeric reporter constructs used in this study were cloned in frame into ptdTomato-C1 expression vector (Clontech Laboratories). All constructs were verified by restriction enzyme digest followed by sequence analysis. Schematic representations of the chimeric ptdTomato constructs used in this study are shown in Figure 6C. The sense and antisense strands of oligonucleotides coding for the stop codon followed by either full-length rat TH 3′UTR (Refseq ID NM__012740.3, nucleotides 1509–1770) or truncated fragments of the TH 3′UTR were commercially synthesized (Life Technologies). Oligonucleotides were annealed and ligated into the BspEI and XbaI sites directly following the ptdTomato ORF. Plasmids were tested for expression of tdTomato by transfecting naive SHY5Y cultures using DNA-In Neuro (GlobalStem) before being used in SCG neuron transfection studies. TH mRNA was synthesized by first inserting DNA oligonucleotides encoding the full length rat TH ORF and 3′UTR (Integrated DNA Technologies) after the T7-promoter of the pBLUE vector using the ECORI and HindIII restriction sites. Following vector linearization, using the HindIII site, 5′ methyl cap containing TH mRNA was synthesized using the mMESSAGE mMACHINE T7 Transcription Kit (Ambion). Ethanol purified TH mRNA (0.5 µg) was transfected into the lateral compartment of Campenot chambers harboring SCG axons using mRNA-In Neuro (GlobalStem).
For characterization of the chimeric mRNA distribution in axons grown in Campenot cultures, DIV 6 SCG cell somas (center compartment) were transfected in 150 µL of Opti-MEM (LifeTechnologies) using 12 µL of DNA-In Neuro (GlobalStem) and 10 µg of desired plasmid DNA in 1.5 mL of neurobasal media. Half the medium was replaced 12–16 h after transfection.
For each experiment, axons expressing the reporter mRNA were imaged using an EVOS digital inverted microscope at 20× magnification. Fluorescence intensity was quantitated using ImageJ software (National Institutes of Health) as described in McCloy et al. (2014) and Burgess et al. (2010). Briefly, axon bundles of interest were selected and measured. The background fluorescence was measured by selecting several regions adjacent to the axon bundles, and the mean background fluorescence intensity was subtracted from that of the measured reporter mRNA signal.
PCR analysis of tdTomato mRNA expression
Distal axons and somas in Campenot chambers were harvested separately, lysed in TRIzol reagent, and RNA isolated using Direct-Zol RNA MiniPrep (Zymo Reasearch) according to the manufacturer's instructions. The RNA samples were eluted in 40 µL of water. Reverse transcription was performed as described above. For qRT-PCR analysis, 4 µL of cDNA (8 ng distal axon RNA, 32 ng for cell body and proximal axon RNA) was added to 10 µL TaqmanFast Universal PCR Master Mix and 1 µL of Custom tdTomato primers and probe (Taqman). For each experimental sample, the reaction was run in triplicate, using a StepOne Real-Time PCR system (Applied Biosystems). The results were analyzed using the StepOne Software (Applied Biosystems), normalizing target mRNA levels to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels. The samples were subsequently electrophoresed on 2.0% Agarose E-Gels (Invitrogen). Taqman probe CCGGTGCCATGCCCCA; forward primer GCCGCCACCACCTGTT; reverse primer CGCTGCCGGTGCTG.
Bioinformatics and statistical analysis
Chimeric gene constructs, oligonucleotides, and primers used in this report were designed using VectorNTI (Invitrogen). The quantitative analysis of cellular fluorescence signals following reporter transfection and immunocytochemistry was performed using ImageJ software as described in McCloy et al. (2014) and Burgess et al. (2010). Multiple sequence alignments were performed using ClustalW, and the secondary structure prediction analysis of the TH 3′UTR was conducted using Mfold (Zuker 2003) and RNA-fold (Gruber et al. 2008). Quantitative data are presented as the mean ± SEM. Student's t-test was used for two-sample comparisons. Analysis of variance (ANOVA) was used to analyze differences among multiple experimental groups and the statistical significance between two groups of a multiple group experiment was determined by Bonferroni corrected post-hoc t-test. Statistical significance was set at α = 0.05 for all experiments. All statistical analyses were performed using Microsoft Excel.
ACKNOWLEDGMENTS
We thank Ms. Cai Chen for invaluable technical assistance. We thank Dr. Lijin Dong (National Institutes of Health/National Eye Institute) for providing us with the pBLUE plasmid. This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health (MH002768).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.053272.115.
REFERENCES
- Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. 2010. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol Cell Neurosci 43: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Kar AN, Natera-Naranjo O, Macgibeny MA, Gioio AE, Kaplan BB. 2012. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci 69: 4017–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod J. 1974. Regulation of the neurotransmitter norepinephrine. In The neurosciences: third study program (ed. Schmitt FO, Wordan FG), pp. 863–876. MIT Press, Cambridge, MA. [Google Scholar]
- Bassell GJ, Singer RH. 1997. mRNA and cytoskeletal filaments. Curr Opin Cell Biol 9: 109–115. [DOI] [PubMed] [Google Scholar]
- Bevilacqua PC, Blose JM. 2008. Structures, kinetics, thermodynamics and biological functions of RNA hairpins. Annu Rev Phys Chem 59: 79–103. [DOI] [PubMed] [Google Scholar]
- Burgess A, Vigneron S, Brioudes E, Labbé J-C, Lorca T, Castro A. 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci 107: 12564–12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM. 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–336. [DOI] [PubMed] [Google Scholar]
- Crispino M, Castigli E, Perrone Capano C, Martin R, Menichini E, Kaplan BB, Giuditta A. 1993. Protein synthesis in a synaptosomal fraction from squid brain. Mol Cell Neurosci 4: 366–374. [DOI] [PubMed] [Google Scholar]
- Crispino M, Kaplan BB, Martin R, Alvarez J, Chun JT, Benech JC, Giuditta A. 1997. Active polysomes are present in the large presynaptic endings of the synaptosomal fraction from squid brain. J Neurosci 17: 7694–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino M, Chun JT, Cefaliello C, Perrone Capano C, Giuditta A. 2014. Local gene expression in nerve endings. Devel Neurobio 74: 279–291. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci 103: 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA. 2011. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 30: 3540–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway MP, Wolf ME, Roth RH. 1986. Regulation of dopamine synthesis in the medial prefrontal cortex is mediated by release modulating autoreceptors: studies in vivo. J Pharmacol Exp Ther 236: 689–698. [PubMed] [Google Scholar]
- Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB. 1991. Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res 28: 18–28. [DOI] [PubMed] [Google Scholar]
- Goetze B, Grunewald B, Kiebler MA, Macchi P. 2003. Coupling the iron-responsive element to GFP—an inducible system to study translation in a single living cell. Sci STKE 204: PL12. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. 2007. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol 27: 701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. 2013. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, DeRisi JL. 2007. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA 13: 625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrott B, Geffen LB. 1972. Rapid axoplasmic transport of tyrosine hydroxylase in relation to other cytoplasmic constituents. Proc Natl Acad Sci 69: 3440–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. 2009. Axonal protein synthesis and the regulation of local mitochondrial function. Results Probl Cell Differ 48: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Kar AN, Gioio AE, Aschrafi A. 2013. MicroRNAs into the axon and presynaptic nerve terminal. Front Cell Neurosci 7: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. 2006. Neuronal RNA granules: movers and makers. Neuron 6: 685–690. [DOI] [PubMed] [Google Scholar]
- Kozicz T. 2001. Axon terminals containing tyrosine hydroxylase- and dopamine-β-hydroxylase immunoreactivity form synapses with galanin immunoreactive neurons in the lateral division of the bed nucleus of the stria terminalis in the rat. Brain Res 914: 23–33. [DOI] [PubMed] [Google Scholar]
- Lenartowski R, Goc A. 2011. Epigenetic, transcriptional and posttranscriptional regulation of the tyrosine hydroxylase gene. Int J Dev Neurosci 29: 873–883. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck F, Lin AC, Allison R. 2006. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci 9: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Melchitzky DS, Gioio A, Solomon Z, Kaplan BB. 1991. Neuronal localization of tyrosine hydroxylase gene products in human neocortex. Molecular Cellular Neurosci 2: 228–234. [DOI] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. 2006. RNA trafficking and local protein synthesis in dendrites, an overview. J Neurosci 26: 7131–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer EJ, Wang DO, Kim S, Barr I, Guo F, Martin KC. 2012. Identification of a cis-acting element that localizes mRNA to synapses. Proc Natl Acad Sci 109: 4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia KR, Trembleau A, Oddi R, Sanna PP, Bloom FE. 1994. Detection and regulation of tyrosine hydroxylase mRNA in catecholaminergic terminal fields: possible axonal compartmentalization. Exp Neurol 130: 394–406. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Nimmrich V, Hernandez AI, Tcherepanov A, Sacktor TC, Tiedge H. 2004. Dendritic transport and localization of protein kinase Mzeta mRNA: implications for molecular memory consolidation. J Biol Chem 279: 52613–52622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. 2010. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16: 1516–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, MacGibeny MA, Gioio AE, Kaplan BB. 2012. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci 49: 263–270. [DOI] [PubMed] [Google Scholar]
- Scott SS, Gervasi NM, Kaplan BB. 2015. Subcellular compartmentalization of neuronal RNAs: an overview. Trends Cell Mol Biol 10: 1–36. [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]