FIGURE 1.
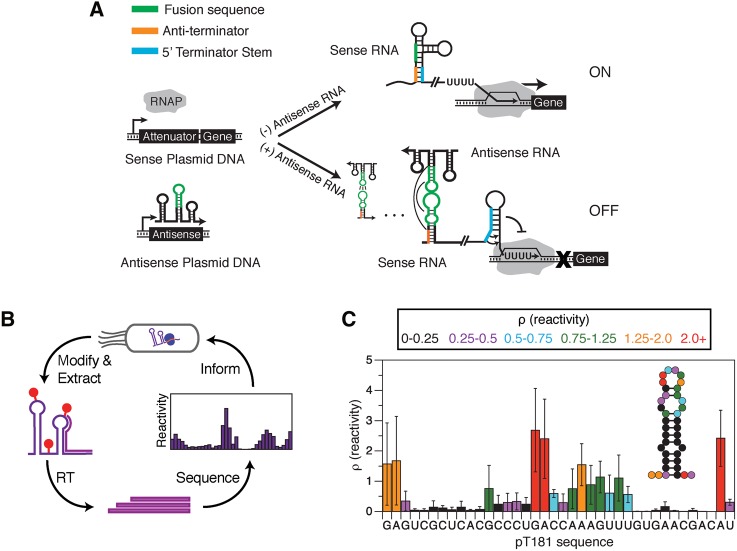
Using in-cell SHAPE-Seq to uncover structure–function design principles for chimeric RNA transcriptional attenuators. (A) Regulation of transcription by chimeric attenuators. Chimeric transcriptional attenuators are engineered by replacing portions of the S. aureus pT181 transcriptional attenuator (Novick et al. 1989; Brantl and Wagner 2000) with RNA-binding regions (fusion sequences) from natural antisense-RNA translational regulators (Takahashi and Lucks 2013). In the absence of antisense RNA, the anti-terminator sequence sequesters the 5′ portion of the terminator stem, preventing terminator formation and allowing transcription elongation by RNA polymerase (RNAP, ON). When antisense RNA is present, its interaction with the attenuator sequesters the anti-terminator, thus allowing terminator formation and preventing downstream transcription (OFF). Antisense/attenuator binding initiates as a kissing hairpin interaction that proceeds to a more extensively paired state, shown schematically with interaction lines (Brantl and Wagner 2000). (B) In-cell SHAPE-Seq (Watters et al. 2016) overview. In-cell SHAPE-Seq characterizes cellular RNA structures using a SHAPE chemical probe that preferentially modifies nucleotides in flexible regions of the RNA. After modification in culture, RNA is extracted, followed by reverse transcription (RT), next-generation sequencing, and bioinformatics steps that yield information about cellular RNA structures in the form of SHAPE reactivity profiles. (C) In-cell SHAPE-Seq reactivity profile for the sensing hairpin of the pT181 attenuator in E. coli. Reactivity profile and restrained secondary structure prediction is shown for the first hairpin of the pT181 attenuator. Color-coded reactivity spectrum represents an average of three independent in-cell SHAPE-Seq experiments, with error bars representing standard deviations at each nucleotide. High reactivities indicate unpaired or unrestrained nucleotides. The hairpin structure (inset) represents the minimum free energy structure generated by RNAstructure (Reuter and Mathews 2010) using average in-cell SHAPE-Seq reactivity data as restraints.