FIGURE 4.
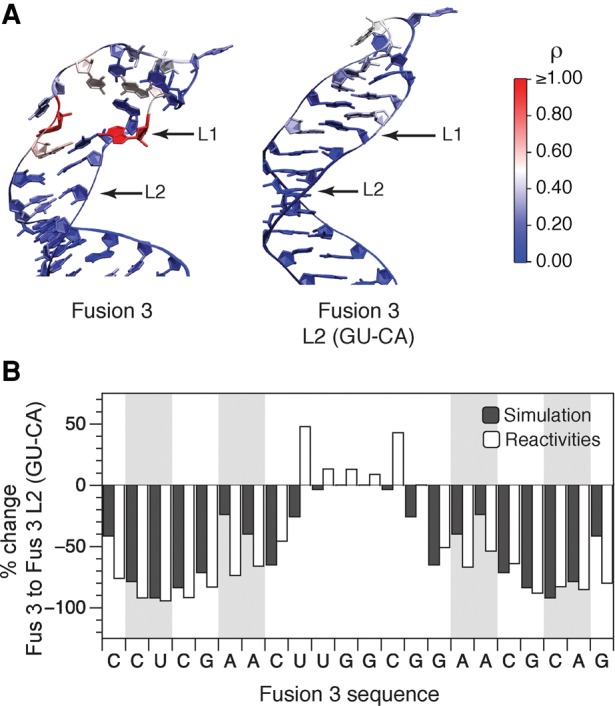
Comparison of in-cell SHAPE-Seq reactivities with molecular dynamics simulations suggests that hairpin interior loops confer structural flexibility. (A) Representative simulation structures of Fusion 3 and Fusion 3 L2(GU–CA). Nucleotides are overlaid with in-cell SHAPE-Seq reactivities from Figure 3F. (B) A comparison of percent change of simulated base-pair opening frequencies and in-cell SHAPE-Seq reactivities between Fusion 3 and Fusion 3 L2(GU–CA) reveals an increase in hairpin structural rigidity (drop in reactivities/opening frequencies) caused by interior loop closing mutations. The 3DNA base-pair search algorithm (Lu and Olson 2003) was used to calculate the percentage of frames in which each base pair in the hairpin was formed over 100 nsec for replica exchange molecular dynamics simulations of Fusion 3 and Fusion 3 L2(GU–CA) (Supplemental Fig. S8). This was then converted into percentages of frames in which each base pair was open (not occupied). This was then used to calculate a percent change in this value from Fusion 3 to Fusion 3 L2(GU–CA) and compared to a percent change in observed in-cell SHAPE-Seq reactivities for each nucleotide. Shaded regions represent nucleotides in the interior loops.
